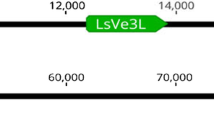
Abstract
Presence of the dominant Tu gene in Lactuca sativa is sufficient to confer resistance to infection by turnip mosaic virus (TuMV). In order to obtain an immunological assay for the presence of TuMV in inoculated plants, the TuMV coat protein (CP) gene was cloned by amplification of a cDNA corresponding to the viral genome using degenerate primers designed from conserved potyvirus CP sequences. The TuMV CP was overexpressed in Escherichia coli, and polyclonal antibodies were produced. To locate Tu on the L. sativa genetic map, F3 families from a cross between cvs “Cobbham Green” (resistant to TuMV) and “Calmar” (susceptible) were genotyped for Tu. Families known to be recombinant in the region containing Tu were infected with TuMV and tested by the indirect enzyme-linked immunosorbent assay (ELISA) using the anti-CP serum. This assay placed Tu between two random amplified polymorphic DNA (RAPD) markers and 3.2 cM from Dm5/8 (which confers resistance to Bremia lactucae). Also, bulked segregant analysis was used to screen for additional RAPD markers tightly linked to the Tu locus. Five new markers linked to Tu were identified in this region, and their location on the genetic map was determined using informative recombinants in the region. Six markers were identified as being linked within 2.5 cM of Tu.
Similar content being viewed by others
References
Abo El-Nil MM, Zettler FW, Hiebert E (1977) Purification, serology and some physical properties of dasheen mosaic virus. Etiology 67:1445–1450
Adams LD (1989) Two-dimensional gel electrophoresis using the ISO-DALT system. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. J. Wiley & Sons, New York, pp 10.3.1–10.3.11
Bowles DJ (1990) Defense-related proteins in higher plants. Annu Rev Biochem 59:873–907
Bruening G, Ponz F, Glascock C, Russel ML, Rowhani A, Chay C (1987) Resistance of cowpeas to Cowpea Mosaic Virus and to Tobacco Ringspot Virus. In: Everet D, Harnett S (eds) Plant resistance to viruses. (Ciba Foundation Symposium 133). J. Wiley & Sons, Chichester, pp 23–32
Choi LK, Wakimoto S (1979) Characterization of the protein components of the turnip mosaic virus. Ann Phytopathol Soc Jpn 45:32–39
Choi JK, Maeda T, Wakimoto S (1977) An improved method for purification of turnip mosaic virus. Ann Phytopathol Soc Jpn 43:440–448
Clark MF, Lister RM, Bar-Joseph M (1986) ELISA techniques. Method Enzymol 118:742–766
Dooner HK (1986) Genetic fine structure of the bronze locus in maize. Genetics 113:1021–1036
Doyle JJ, Doyle JL (1987) Isolation of plant DNA from fresh tissue. Phytochem Bull 19:11–14
Duffus J, Zink FW (1969) A diagnostic host reaction for the identification of turnip mosaic virus. Plant Dis Rep 53:916–917
Ellis JG, Lawrence GJ, Peacock WJ, Pryor AJ (1988) Approaches to cloning plant genes conferring resistance to fungal pathogens. Annu Rev Phytopathol 26:245–263
Fraser RSS (1987) Biochemistry of virus-infected plants. J. Wiley & Sons, New York
Fraser RSS (1990) The genetics of resistance to plant viruses. Annu Rev Phytopathol 28:179–200
Gallagher S, Winston SE, Fuller SA, Hurrell JGR (1989) Immunoblotting and Immunodetection. In: Ausubel FM. Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. J. Wiley & Sons, New York, pp 10.8.1–10.8.14
Ganal MW, Young ND, Tanksley SD (1989) Pulsed field gel electrophoresis and physical mapping of large DNA fragments in the Tm-2a region of chromosome 9 in tomato. Mol Gen Genet 215:396–400
Harlow E, Lane D (1988) Isolation of protein antigens from bacterial overexpression systems: inclusion bodies. In: Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., p 726
Hiebert E, McDonald JG (1976) Capsid protein heterogeneity in turnip mosaic virus. Virology 70:144–150
Kesseli RV, Paran I, Ochoa O, Wang WC, Michelmore RW (1993) Linkage map of lettuce (Lactuca sativa). In: O'Brien SJ (ed) Genetic maps, vol 6. Cold Spring Harbor Press, Cold Spring Harbor, N.Y., pp 229–233
Kombrink E, Schroder M, Hahlbrock K (1988) Several “pathogenesis-related” proteins in potato are 1,2-β-glucanases and chitinases. Proc Natl Acad Sci USA 85:782–786
Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, Newburg L (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:171–181
Legrand M, Kaufmann S, Geoffroy P, Fritig B (1987) Biological function of-pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Natl Acad Sci USA 84:6750–6754
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Michelmore RW, Kesseli RV, Francis DM, Paran I, Fortin MG, Yang C-H (1992) Strategies for cloning plants disease resistance genes. In: Gurr SJ, McPherson MJ, Bowles DJ (eds) Molecular plant pathology, vol 2: a practical approach. IRL Press, Oxford, pp 231–288
Nicolas O, Laliberté J-F (1992) The complete nucleotide sequence of turnip mosaic virus RNA. J Gen Virol 73:2785–2793
Ryder EJ (1979) Vanguard 75 lettuce. HortScience 14:284–286
Studier FM, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Method Enzymol 185:60–89
Tremblay MF, Nicolas O, Sinha RC, Lazure C, Laliberté J-F (1990) Sequence of the 3′-terminal region of turnip mosaic virus RNA and the capsid protein gene. J Gen Virol 71:2769–2772
Vaitukaitis JL (1981) Production of antisera with small doses of immunogen: multiple interdermal injections. Method Enzymol 73:46–52
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Zink FW, Duffus JE (1970) Linkage of turnip mosaic virus susceptibility and downy mildew, Bremia lactucae, resistance in lettuce. J Am Soc HortSci 95:420–422
Author information
Authors and Affiliations
Additional information
Communicated by G. E. Hart
Rights and permissions
About this article
Cite this article
Robbins, M.A., Witsenboer, H., Michelmore, R.W. et al. Genetic mapping of turnip mosaic virus resistance in Lactuca sativa . Theoret. Appl. Genetics 89, 583–589 (1994). https://doi.org/10.1007/BF00222452
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00222452