Abstract
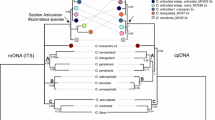
Phylogenetic relationships of the Poaceae subfamily, Pooideae, were estimated from the sequences of the internal transcribed spacer (ITS) region of nuclear ribosomal DNA. The entire ITS region of 25 species belonging to 19 genera representing seven tribes was directly sequenced from polymerase chain reaction (PCR)-amplified DNA fragments. The published sequence of rice, Oryza saliva, was used as the outgroup. Sequences of these taxa were analyzed with maximum parsimony (PAUP) and the neighbor-joining distance method (NJ). Among the tribes, the Stipeae, Meliceae and Brachypodieae, all with small chromosomes and a basic number more than x=7, diverged in succession. The Poeae, Aveneae, Bromeae and Triticeae, with large chromosomes and a basic number of x=7, form a monophyletic clade. The Poeae and Aveneae are the sister group of the Bromeae and Triticeae. On the ITS tree, the Brachypodieae is distantly related to the Triticeae and Bromeae, which differs from the phylogenies based on restriction-site variation of cpDNA and morphological characters. The phylogenetic relationships of the seven pooid tribes inferred from the ITS sequences are highly concordant with the cytogenetic evidence that the reduction in chromosome number and the increase in chromosome size evolved only once in the pooids and pre-dated the divergence of the Poeae, Aveneae, Bromeae and Triticeae.
This paper reports factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable
Similar content being viewed by others
References
Avdulov NP (1931) Karyo-systematische Untersuchungen der Familie Gramineen. Bull Appl Bot Genet Plant Breed Suppl 44:1–428
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants:an example from the Compositae. Mol Phylog Evol 1:3–16
Baldwin BG (1993) Molecular phylogenetics of Calycadenia (Compositae) based on ITS sequences of nuclear ribosomal DNA:chromosomal and morphological evolution reexamined. Am J Bot 80:222–238
Barkworth ME (1982) Embryological characters and the taxonomy of the Stipeae (Gramineae). Taxon 31:233–243
Barkworth ME, Everett J (1987) Evolution in the Stipeae identification and relationships of its monophyletic taxa. In:Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Institution, Washington D. C., pp 251–264
Baum BR (1987) Numerical taxonomic analyses of the Poaceae. In:Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Instituation, Washington, D. C., pp 334–342
Clayton WD (1981) Evolution and distribution of grasses. Ann Mis Bot Gard 68:5–14
Clayton WD, Renvoize SA (1986) Genera graminum. Kew Bull Add Ser 13:1–389
Davis JI, Soreng RJ (1993) Phylogenetic structure in the grass family (Poaceae) as inferred from chloroplast DNA restriction-site variation. Am J Bot 80:1444–1454
Dewey DR (1982) Genomic and phylogenetic relationships among North American perennial Triticeae grasses. In:Estes JR, Tyrl RJ, Brunken JN (eds) Grasses and grasslands systematics and ecology. University of Oklahoma Press, Oklahoma, pp 51–80
Dewey DR (1984) The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In:Gustafson JP (ed) Gene manipulation in plant inprovement. Plenum Publishing Corporation, New York, pp 209–279
Donoghue MJ, Sanderson MJ (1992) The suitability of molecular and morphological evidence in reconstructing plant phytogeny. In:Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York London, pp 340–368
Enomoto SI, Ogihara Y, Tsunewaki K (1985) Studies on the origin of crop species by restriction endonuclease analysis of organellar DNA. I. Phylogenetic relationships among ten cereals revealed by the restriction fragment patterns of chloroplast DNA. Jpn J Genet 60:411–424
Flavell RB (1982) Sequence amplification, deletion and rearrangement:major sources of variation during species divergence. In:Dover GA, Flavell RB (eds) Genome evolution. Academic Press Eondon, pp 301–323
Gould FW, Shaw RB (1983) Grass systematics, 2nd edn. Texas A and M University press, College Station, Texas
Higgins DG, Bleasby AJ, Fuchs R (1992) CLUSTAL V:Improved Software for multiple sequence alignment. CABIOS 8:189–191
Hilu KW, Wright K (1982) Systematics of Gramineae:a cluster analysis study. Taxon 31:9–36
Hsiao C, Chatterton NJ, Asay KH, Jensen KB (1994) Phylogenetic relationships of 10 grass species:an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome 37:112–120
Hsiao C, Chatterton NJ, Asay KH, Jensen KB (1995) Phylogenetic relationships of the monogenomic species of the wheat tribe, Triticeae (Poaceae), inferred from nuclear rDNA (ITS) sequences. Genome (in press)
Kellogg EA (1992) Tools for studying the chloroplast genome in the Triticeae (Gramineae):an EcoRI map, a diagnostic deletion, and support for Bromus as an outgroup. Am J Bot 79:186–197
Kellogg EA, Campbell CS (1987) Phylogenetic analyses of the Gramineae. In:Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Institution, Washington, pp 310–322
Kellogg EA, Watson L (1993) Phylogenetic studies of a large data set. I. Bambusoideae, Andropogonodae, and Pooideae (Gramineae). Bot Rev 59:273–343
Kihara H (1954) Considerations on the evolution and distribution of Aegilops species based on the analyzer method. Cytologia 19:336–357
Kumar S, Tamura K, Nei M (1993) MEGA, Molecular Evolutionary Genetics Analysis, version 1.0. Institute of Molecular Evolutionary Genetics, The Pennsylvania State University, University Park, Pennsylvania
Lassner MW, Peterson P, Yoder JI (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7:116–128
Macfarlane TD (1987) Poaceae subfamily Pooideae. In:Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Institution, Washington D. C., pp 265–276
Macfarlane TD, Watson L (1980) The circumscription of Poaceae subfamily Pooideae, with notes on some controversial genera. Taxon 29:645–666
Macfarlane TD, Watson E (1982) The classification of Poaceae subfamily Pooideae. Taxon 31:178–203
Sharma ML (1979) Some considerations on the phylogeny and chromosomal evolution in grasses. Cytologia 44:679–685
Smith D (1973) The nonstructural carbohydrates. In:Butler GW, Bailey RW (eds) Chemistry and biochemistry of herbage. Academic Press, Eondon New York 1:105–155
Soreng RJ, Davis JI, Doyle JJ (1990) A phylogenetic analysis of chloroplast DNA restriction site variation in Poaceae subfam. Pooideae. Plant Syst Evol 172:83–97
Stebbins GL (1956) Cytogenetics and evolution of the grass family. Am J Bot 43:890–905
Stebbins GL (1982) Major trends of evolution in the Poaceae and their possible significance. In:Estes JR, Tyrl RJ, Brunken JN (eds) Grasses and grasslands. University of Oklahoma Press, Oklahoma, pp 3–36
Stebbins GL (1987) Grass systematics and evolution:past, present and future. In:Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution, Smithsonian Institute, Washington, D. C., pp 359–367
Stebbins GE, Crampton B (1961) A suggested revision of the grass genera of temperate north America. Rec Adv Bot 1:133–145
Suh Y, Thien EB, Reeve HE, Zimmer EA (1993) Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of ribosomal DNA in Winteraceae. Am J Bot 80:1042–1055
Swofford DE (1993) PAUP, phylogenetic analysis using parsimony. Version 3.1.1. for Apple Macintosh. Illinois Natural History Survey, Champaign, Illinois
Takaiwa F, Oono K, Sugiura M (1985) Nucleotide sequence of the 17s–25s spacer region from rice rDNA. Plant Mol Biol 4:355–364
Tateoka T (1957) Miscellaneous papers on the phylogeny of Poaceae (10). Proposition of a new phylogenetic system of Poaceae. J Jap Bot 32:275–287
Tateoka T (1962) Starch grains of endosperm in grass systematics. Bot Mag (Tokyo) 75:377–383
Tzvelev NN (1989) The system of grasses (Poaceae) and their evolution. Bot Rev 55:141–204
Watson L, Knox RB (1976) Pollen wall antigens and allergens:taxonomically-ordered variation among grasses. Ann Bot 40:399–408
Watson L, Clifford HT, Dallwitz MJ (1985) The classification of Poaceae:subfamilies and supertribes. Aust J Bot 33:433–484
Williams JGK, Hanafey MK, Rafalski JA, Tingey SV (1993) Genetic analysis using random amplified polymorphic DNA markers. Methods Enzymol 218:704–740
Wojciechowski MF, Sanderson MJ, Baldwin BG, Donoghue MJ (1993) Monophyly of aneuploid astragalus (Fabaceae):evidence from nuclear ribosomal DNA internal transcribed spacer sequences. Am J Bot 80:711–722
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, choloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Zurawski G, Clegg MT (1987) Evolution of higher plant chloroplast DNA-encoded genes implications for structure-function and phylogenetic studies. Annu Rev Plant Physiol 38:391–418
Author information
Authors and Affiliations
Additional information
Communicated by Dooner
This paper is a cooperative investigation of USDA-ARS and the Utah Agricultural Experiment Station. Logan, Utah 84322. Journal Paper No. 4581
Rights and permissions
About this article
Cite this article
Hsiao, C., Chatterton, N.J., Asay, K.H. et al. Molecular phylogeny of the Pooideae (Poaceae) based on nuclear rDNA (ITS) sequences. Theoret. Appl. Genetics 90, 389–398 (1995). https://doi.org/10.1007/BF00221981
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00221981