Summary
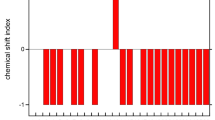
Melittin is a naturally occurring hexacosa peptide which forms an amphiphilic helix in methanol, a random coil in water, and a tetramer of helices at basic pH or in the presence of a high salt concentration. The monomeric structure in methanol has been well characterized by proton NMR (Pastore et al. (1989) Eur. Biophys. J., 16, 363–367). In the present paper, chemical shifts of the backbone α-carbons of melittin in methanol were determined by mapping previously published α-proton shifts (Bazzo et al. (1988) Eur. J. Biochem., 173, 139–146), to natural abundance {1H}13C cross peaks appearing in the 2D heteronuclear multiple-quantum NMR spectrum. Changes in chemical shifts consequent to stepwise increases in the percentage of water in a mixed methanol/water solvent system were observed in similar spectra. The α-carbon shifts varied more smoothly than the corresponding α-proton shifts and were found to correlate with the transition from the helix to the random coil conformer in parallel with changes in the circular dichroism spectrum. Chemical shifts of this peptide are interpreted with regard to the current database of assignments in proteins of known 3D structure (Wishart et al. (1991) J. Mol. Biol., 222, 311–333). The N-terminal region of the peptide shows increased flexibility at lower methanol concentrations, as evidenced by the merger of the α-proton resonances of G1 (at 40% and 15% methanol) and G3 (at 15% methanol). Conformational exchange rates for G1 and G3 were estimated by comparison of the experimental spectra with simulated spectra and found to be as large as 4000 s−1 for G1 in 40% and 15% methanol and 600 s−1 for G3 in 15% methanol. Overall, these 1H and 13C chemical shift data support the description of monomeric melittin in methanol currently evolving in the literature and suggest a structure composed of a linked pair of helices with different structural stabilities, each of which experiences dynamical fraying at its free terminus.
Similar content being viewed by others
References
Bazzo R., Tappin M.J., Pastore A., Harvey T.S., Carver J.A. and Campbell I.D. (1988) Eur. J. Biochem., 173, 139–146.
Bello J., Bello H.R. and Granados E. (1982) Biochemistry, 21, 461–465.
Bax A., Griffey R.H. and Hawkins B.L. (1983) J. Magn. Reson., 55, 301–315.
Braunschweiler L. and Ernst R.R. (1983) J. Magn. Reson., 53, 521–528.
Brown L.R., Lauterwein J. and Wüthrich K. (1980) Biochem. Biophys. Acta., 62, 231–244.
Buckley P.J., Kemple M.D. and Prendergast F.G. (1993) J. Cell. Biochem., 17C, 291.
Dempsey C.E. (1988) Biochemistry, 27, 6893–6901.
Kaplan J.I. and Fraenkel G. (1980) NMR of Chemically Exchanging Systems, Academic Press, New York.
Gans P.J., Lyu P.C., Manning M.C., Woody R.W. and Kallenbach N.R. (1991) Biopolymers, 31, 1605–1614.
Greenfield N. and Fasman G.D. (1969) Biochemistry, 8, 4108–4116.
Habermann E. (1972) Science, 177, 314–322.
Ikura T., Gō N. and Inagaki F. (1991) Proteins: Struct. Func. Genet., 9, 81–89.
JohnsonJr. W.C. (1990) Proteins: Struct. Func. Genet., 7, 205–214.
Lakowicz J.R., Gryczynski I., Wiczk W., Laczko G., Prendergast F.G. and Johnson M. (1990) Biophys. Chem., 36, 99–115.
Lauterwein J., Brown L.R. and Wüthrich K. (1980) Biochem. Biophys. Acta., 622, 219–230.
Liff M.I., Lyu P.C. and Kallenbach N.R. (1991) J. Am. Chem. Soc., 113, 1014–1019.
Merutka G., Shalongo W. and Stellwagen E. (1991) Biochemistry, 30, 4245–4248.
Müller L. (1979) J. Am. Chem. Soc., 101, 4481–4484.
Pastore A., Harvey T.S., Dempsey C.E. and Campbell I.D. (1989) Eur. Biophys. J., 16, 363–367.
Pastore A. and Saudek V. (1990) J. Magn. Reson., 90, 165–176.
Quay S.C. and Condie C.C. (1983) Biochemistry, 22, 695–700.
Reily M.D., Venkataraman T. and Omecinsky D.O. (1992) J. Am. Chem. Soc., 114, 6251–6252.
Schwarz G. J. Mol. Biol. (1965), 11, 64–77.
Spera S. and Bax A. J. Am. Chem. Soc. (1991), 113, 5490–5492.
Terwillinger T.C. and Eisenberg D. (1982a) J. Biol. Chem., 257, 6010–6015.
Terwillinger T.C. and Eisenberg D. (1982b) J. Biol. Chem., 257, 6016–6022.
Wesson L. and Eisenberg D. (1992) Protein Sci., 1, 227–235.
Weaver A.J., Kemple M.D., Brauner J.W., Mendelson R. and Prendergast F.G. (1992) Biochemistry, 28, 1301–1313.
Weaver A.J., Kemple M.D. and Prendergast F.G. (1989) Biochemistry, 28, 8614–8623.
Wilcox W. and Eisenberg D. (1992) Protein Sci., 1, 641–653.
Wishart D.S., Sykes B.D. and Richards F.M. (1991) J. Mol. Biol., 222, 311–333.
Yunes R.A. (1982) Arch. Biochem. Biophys., 216, 559–565.
Zhou N.E., Zhu B., Sykes B.D. and Hodges R.S. (1992) J. Am. Chem. Soc., 114, 4320–4326.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Buckley, P., Edison, A.S., Kemple, M.D. et al. 13Cα-NMR assignments of melittin in methanol and chemical shift correlations with secondary structure. J Biomol NMR 3, 639–652 (1993). https://doi.org/10.1007/BF00198369
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00198369