Abstract
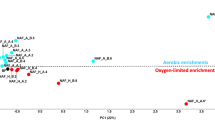
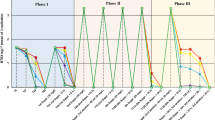
In the course of an in situ bioremediation, different hydrologically controllable test plots were installed on the ground of a waste-oil contaminated site, and continuously injected with nutrient solution and the electron acceptors NO3 −, O2, and H2O2. In a two-year period, groundwater samples obtained from different recovery wells within these field plots, in addition to subsoil samples, were monitored for several chemical and microbiological parameters. The removal of hydrocarbons observed in the water samples could not unambiguously be attributed to biodegradation, and was probably caused by groundwater treatment measures. However, chemical (gaschromatographic) and microbiological data from the subsoil samples indicated a biological degradation of pollutants. Analysis of the groundwater samples of the different test plots revealed only minor quantitative differences. With time, only a slight increase in bacterial numbers on different media, including hydrocarbon-agar, was observed. In general, chemical and microbiological analyses of groundwater samples cannot replace analyses of subsoil samples for a sufficient documentation of in situ remediation processes in subsoil. From the groundwater and subsoil samples, 3,446 pure cultures, obtained from R2A agar, were characterized morphologically and physiologically, and identified in order to study the culturable bacterial communities. Several qualitative differences in composition and diversity of the bacterial communities among the test plots were observed. More than 70 different species or taxonomic groups (most of them known as hydrocarbon degrading taxa) could be identified from the groundwater samples; these were mainly the Gram-negative genera Acinetobacter, Alcaligenes, Comamonas, Hydrogenophaga, Pseudomonas, Flavobacterium/Flexibacter/Cytophaga, and others. A high proportion of Gram-positive organisms (42.5%), belonging to Bacillus and the various genera of coryneform and nocardioform organisms, were isolated from the subsoil samples.
Similar content being viewed by others
References
Amann RI, Binder BJ, Olson RJ, Chrisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Amann RI, Krumholz L, Stahl DA (1990) Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770
Arendt F, Hinsenveld M, van den Brink WJ (eds) (1990) Altlastensanierung '90. Kluwer Academic Publishers, Dordrecht, Boston
Atlas RM (1981) Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microb Rev 45:180–209.
Atlas RM (1984) Diversity of microbial communities, In: Marshall KC (ed) Advances in microbial ecology. Plenum Press, New York, pp 1–47
Austin B, Garges S, Conrad B, Harding EE, Colwell RR, Simudu U, Taga N (1979) Comparative study of aerobic, heterotrophic bacteria flora of Chesapeake Bay and Tokyo Bay. Appl Environ Microbiol 37:704–714
Balkwill DL, Ghiorse WC (1985) Characterization of subsurface bacteria associated with two shallow aquifers in Oklahoma. Appl Environ Microbiol 50:580–588
Battermann, G (1986) Decontamination of polluted aquifers by biodegradation. In: Assink JW, van den Brink WJ (eds) Contaminated soils. Nijhoff Publishers, Dordrecht, pp 711–722
Bianchi MAG, Bianchi AJM (1982) Statistical sampling of bacterial strains and its use in bacterial diversity measurements. Microb Ecol 8:61–69
Brock, TD (1987) The study of microorganisms in situ: progress and problems. In: Fletcher M, Gray TRG, Jones JG (eds) Ecology of microbial communities, Symposium 41. The Society of for General Microbiology. Cambridge University Press, Cambridge, pp 1–17
Buchanan-Mappin JM, Wallis PM, Buchanan AG (1986) Enumeration and identification of heterotrophic bacteria in groundwater and in mountain stream. Can J Microbiol 32:93–98
Busse H-J, El-Banna T, Auling G (1989) Evaluation of different approaches for identification of xenobiotic-degrading pseudomonads. Appl Environ Microbiol 55:1578–1583
Busse H-J, El-Banna T, Oyaizu H, Auling G (1992) Identification of xenobiotic-degrading isolates from the beta subclass of the Proteobacteria by a polyphasic approach including 16S rRNA partial sequencing. Int J Syst Bacteriol 42:19–26
Deutsche Einheitsverfahren zur Wasseruntersuchung, DEV (1971, 1981) Mikrobiologische Verfahren, Gruppe K5:1–8; Summarische Wirkungs- und Stoffkenngröβen, H18:1–10. Verlag Chemie, Weinheim.
De Vos P, De Ley J (1983) Intra- and intergeneric similarities of Pseudomonas and Xanthomonas ribosomal ribonucleic acid cistrons. Int J Syst Bacteriol 33:487–509
Doetsch, RN (1981) Determinative methods in light microscopy. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips EB (eds) Manual of methods for general microbiology. American Society for Microbiology, Washington DC, pp 21–33
Fox GE, Wisotzkey JD, Jurtshuk Jr P (1992) How close is close: 16S rRNA sequence identity may not guarantee species identity. Int J Syst Bacteriol 42:166–170
Frederickson JK, Balkwill DL, Zachara JM, Li SMW, Brockman FJ, Simmons MA (1991) Physiological diversity and distributions of heterotrophic bacteria in deep cretaceous sediments of the Atlantic coastal plain. Appl Environ Microbiol 57:402–411
Gamble TN, Betlach MR, Tiedje JM (1977) Numerically dominant denitrifying bacteria from world soils. Appl Environ Microbiol 33:926–939
Garland JL, Mills AA (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Gassmann G (1981) Chromatographic separation of diasteriomeric isoprenoids for the identification of fossil oil contamination. Mar Pollut Bull 12:78–84
Gehlen M, Trampisch HJ, Dott W (1985) Physiological characterization of heterotrophic bacterial communities from selected aquatic environments. Microb Ecol 11:205–219
Ghiorse WC, Balkwill DL (1983) Enumeration and morphological characterization of bacteria indigenous to subsurface environments. Dev Ind Microbiol 24:213–224
Herbert D, Phipps PJ, Strange RE (1971) Chemical analysis of microbial cells. Meth Microbiol 513:209–344
Holmes B, Hill LR (1985) Computers in diagnostic bacteriology, including identification. In Goodfellow M, Jones D, Priest FG (eds) Computer assisted bacterial systematics. Academic Press, London, pp 265–287
Holmes B, Owen RJ, McMeekin TA (1984) Genus Flavobacterium Bergey, Harrison, Breed, Hammer, and Huntoon 1923 97AL. In Krieg NR, Holt JH (eds) Bergey's manual of systematic bacteriology, vol 1. Williams & Wilkins, Baltimore, pp 353–361
Jones JG (1987) Diversity in freshwater microbiology. In Fletcher M, Gray TRG, Jones JG (eds) Ecology of microbial communities, Symposium 41. The Society for General Microbiology, Cambridge University Press, Cambridge, pp 235–259
Kämpfer P (1988) Automatisierte Charakterisierung mikrobieller Lebensgemeinschaften Hygiene Berlin 1 Veröffentlichungen aus dem Fachgebiet Hygiene der Technischen Universität Berlin und dem Institut für Hygiene der Freien Universität Berlin ISBN 3 7983 1226 5, Berlin
Kämpfer P (1991) Application of miniaturized physiological tests in numerical classification and identification of some bacilli. J Gen Appl Microbiol 37:225–247
Kämpfer P, Dott W (1988) Differentiation of some Gram-negative glucose nonfermenting bacteria using miniaturized carbon sources assimilation tests. Zentralbl Bakt Hyg B186:468–477
Kämpfer P, Dott W, Kroppenstedt RM (1990) Numerical classification and identification of some nocardioform bacteria. J Gen Appl Microbiol 36:309–331
Kampfer P, Steiof M, Dott W (1991) Microbiological characterization of a fuel-oil contaminated site including numerical identification of heterotrophic water and soil bacteria. Microb Ecol 21:227–251
Keddie RM, Collins MD, Jones D (1986) Genus Arthrobacter Conn and Dimmick 1947, 300AL. In Sneath PHA, Mair NS, Sharpe ME, Holt JH (eds) Bergey's manual of systematic bacteriology, vol 2. Williams & Wilkins, Baltimore, pp 1288–1301
Kölbel-Boelke J, Tienken B, Nehrkorn A (1988) Microbial communities in the saturated groundwater environment. I. Methods for isolation and characterization of heterotrophic bacteria. Microb Ecol 16:17–29
Kölbel-Boelke J, Anders EM, Nehrkorn A (1988) Microbial communities in the saturated groundwater environment. II. Diversity of bacterial communities in a pleistocene sand aquifer and their in vitro activities. Microb Ecol 16:31–48
Leahy JG, Colwell RR (1990) Microbial degradation of hydrocarbons in the environment. Microb Rev 54:305–315
Lee MD, Ward CH (1985) Biological methods for the restoration of contaminated aquifers. Environ Toxic Chem 4:743–750
Lee MD, Thomas JM, Borden RC, Bedient PB, Ward CH, Wilson JT (1988) Biorestoration of aquifers contaminated with organic compounds. CRC Crit Rev Environ Contr 18:29–89
Lindstrom JE, Prince RC, Clark JC, Grossman MJ, Yeager TR, Braddock JF, Brown EJ (1991) Microbial populations and hydrocarbon biodegradation potentials in fertilized shoreline sediments affected by the T/V Exxon Valdez oil spills. Appl Environ Microbiol 57:2514–2522
Lipsky A, Klatte S, Bendinger B, Altendorf K (1992) Differentiation of Gram-negative, bacteria isolated from biofilters on the basis of fatty acid composition, quinone system, and physiological reaction profiles. Appl Environ Microbiol 58:2053–2065
Morgan P, Watkinson RJ (1989) Hydrocarbon degradation in soils and methods for soil biotreatment. CRC Crit Rev Biotechnol 8:305–333
Obst U, Holzapfel-Pschorn A (1988) Enzymatische Tests fur die Wasseranalytik. Oldenbourg Verlag, Munich
Olsen RA, Bakken LR (1987) Viability of soil bacteria: optimization of plate-counting technique and comparison between total counts and plate counts within different size groups. Microbial Ecol 13:59–74
Overbeck J, Chrost RJ (ed) 1990 Aquatic microbial ecology. Springer Verlag, Heidelberg, New York
Pace NR, Stahl DA, Lane DJ, Olsen GJ (1986) The analysis of natural microbial populations by ribosomal RNA sequences. In: Marshall KC (ed) Advances in microbial ecology, vol 9. Plenum Press, New York, pp 1–55
Palleroni NJ (1984) Genus I Pseudomouas Migula 1894, 237AL. In Krieg NR, Holt JH (eds) Bergey's manual of systematic bacteriology, vol 1. Williams & Wilkins, Baltimore, pp 141–199
Parkes RJ (1987) Analysis of microbial communities within sediments using biomarkers. In: Fletcher M, Gray TRG, Jones JG (eds) Ecology of microbial communities, Symposium 41. The Society of for General Microbiology, Cambridge University Press, Cambridge, pp 147–177
Pfennig N, Lippert KD (1966) Über das Vitamin B12 Bedürfnis phototropher Schwefelbakterien. Arch Microbiol 55:245–256
Püttmann W (1988) Microbial degradation of petroleum in contaminated soils—analytical aspects. In: Wolf K, van der Brink WJ, Colon FJ (eds) Contaminated soil '88. Kluwer Academic Publishers, Dordrecht, London, pp 189–199
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Ridgway HF, Safarik J, Phipps D, Carl P, Clark D (1990) Identification and catabolic activity of well-derived gasoline-degrading bacteria from a contaminated aquifer. Appl Environ Microbiol 56:3565–3575
Ripper P, Früchtenicht H (1989) Umweltschadensfall Pintsch-Ö1 Hanau: Sanierungsschritte eingeleitet. WLB Wasser Luft Boden 4:60–63
Riss A (1992) Mikrobiologische On-Site und In-Situ-Pilot Versuche zur Sanierung des Altölraffineriestandorts Pintsch, Hanau. In: DECHEMA-Fachgespräche Umweltschutz: Mikrobiologische Reinigung von Böden. DECHEMA, Frankfurt, pp 134–153
Riss A, Barenschee ER, Helmling O, Ripper P (1991) Einsatz von Wasserstoffperoxid zum mikrobiologischen Kohlenwasserstoffabbau. gwf Wasser Abwasser 132:115–126
Rosenberg E(1992) The hydrocarbon-oxidizing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd ed, vol 1, Springer-Verlag, New York, pp 446–459
Sayler GS, Shields MS, Tedford E, Breen A, Hooper S, Sirotkin K, Davis J (1985) Application of DNA-DNA colony hybridization for the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol 49:1295–1303
Smith GA, Nickels JS, Kerger BD, Davis JD, Collins SP (1985) Quantitative characterization of microbial biomass and community structure in subsurface material: a procaryotic consortium responsive to organic contamination. Can J Microbiol 32:104–111
Spring S, Amann R, Ludwig W, Schleifer K-H, Petersen N (1992) Phylogenetic diversity and identification of nonculturable, magnetotactic bacteria. Syst Appl Microbiol 15:116–122
Sneath PHA (1979) BASIC program for identification of an unknown with presence-absence data against an identification matrix of percent positive characters. Comput Geosci 5:195–213
Song HG, Bartha R (1990) Effects of jet fuel spills on the microbial community of soil. Appl Env Microbiol 56:646–652
Steffan RJ, Atlas RM (1991) Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol 45:133–161
Stenström I-M, Molin G (1990) Classification of the spoilage flora of fish, with special reference to Shewanella putrefaciens. J Appl Bacteriol 68:601–618
Weller R, Weller JW, Ward DM (1989) Selective recovery of 16S rRNA sequences from natural microbial communities in the form of cDNA. Appl Environ Microbiol 55:1818–1822
Westlake DWS, Jobson A, Phillippe R, Cook FD (1974) Biodegradability of crude oil composition. Can J Microbiol 20:915–928
Willcox WR, Lapage SP, Bascomb S, Curtis MA (1973) Identification of bacteria by computer: theory and programming. J Gen Microbiol 77:317–330
Willems A, De Vos P, De Ley J (1992) The genus Comamonas. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd ed, vol 3, Springer-Verlag, New York, pp 2583–2590
Wilson IT, McNabb JF, Balkwill DL, Ghiorse WC (1983) Enumeration and characterization of bacteria indigenous to a shallow watertable aquifer. Groundwater 21:134–142
Wilson JT, McNabb JF, Wilson BH, Noonan NJ (1983) Biotransformation of selected organic pollutants in ground water. Dev Ind Microbiol 24:225–233
Zarda B, Amann R, Wallner G, Schleifer K-H (1991) Identification of single bacterial cells using digoxigenin-labelled, rRNA-targeted oligonucleotides. J Gen Microbiol 137:2823–2830
Author information
Authors and Affiliations
Additional information
Offprint requests to: P. Kämpfer.
Rights and permissions
About this article
Cite this article
Kämpfer, P., Steiof, M., Becker, P.M. et al. Characterization of chemoheterotrophic bacteria associated with the in situ bioremediation of a waste-oil contaminated site. Microb Ecol 26, 161–188 (1993). https://doi.org/10.1007/BF00177050
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00177050