Abstract
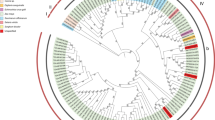
Isolates of cauliflower mosaic virus (CaMV) differ in host range and symptomatology. Knowledge of their sequence relationships should assist in identifying nucleotide sequences responsible for isolate-specific characters. Complete nucleotide sequences of the DNAs of eight isolates of CaMV were aligned and the aligned sequences were used to analyze phylogenetic relationships by maximum likelihood, bootstrapped parsimony, and distance methods. Isolates found in North America clustered separately from those isolated from other parts of the world. Additional isolates, for which partial sequences were available, were incorporated into phylogenetic analysis of the sequences of genome segments corresponding to individual protein coding regions or the large intergenic region of CaMV DNA. The analysis revealed several instances where the position of an isolate on a tree for one coding region did not agree with the position of the isolate on the tree for the complete genome or with its position on trees for other coding regions. Examination of the distribution of shared residue types of phylogenetically informative positions in anomalous regions suggested that most of the anomalies were due to recombination events during the evolution of the isolates. Application of an algorithm that searches for segments of significant length that are identical between pairs of isolates or contain a significantly high concentration of polymorphisms suggested two additional recombination events between progenitors of the isolates studied and an event between the XinJing isolate and a CaMV not represented in the data set. An earlier phylogenetic origin for CaMV than for carnation etched ring virus, the caulimovirus used as outgroup in these analyses, was deduced from the position of the outgroup with North American isolates in some trees, but with non-North American isolates in other trees.
Similar content being viewed by others
References
Balàzs E, Guilley H, Jonard G, Richards K (1982) Nucleotide sequence of DNA from an altered-virulence isolate D/H of the cauliflower mosaic virus. Gene 19:239–249
Blok J, Mackenzie A, Guy P, Gibbs A (1987) Nucleotide sequence comparisons of turnip yellow mosaic virus isolates from Australia and Europe. Arch Virol 97:283–295
Chenault KD, Melcher U (1993) The complete nucleotide sequence of cauliflower mosaic virus isolate BBC. Gene 123:255–257
Chenault KD, Melcher U (1994) Patterns of nucleotide sequence variation among cauliflower mosaic virus isolates. Biochimie 76:3–8
Choe IS, Melcher U, Richards K, Lebeurier G, Essenberg RC (1985) Recombination between mutant cauliflower mosaic virus DNAs. Plant Mol Biol 5:281–289
Daubert SD, Schoelz J, Debao L, Shepherd RJ (1984) Expression of disease symptoms in cauliflower mosaic virus genomic hybrids. J Mol Appl Gen 2:537–547
Dawson WO (1992) Tobamovirus-plant interactions. Virology 186: 359–367
Dixon L, Nyffenegger T, Delley G, Martinez-Izquierdo J, Hohn T (1986) Evidence for replicative recombination in cauliflower mosaic virus. Virology 150:463–468
Donis R, Bean WJ, Kawaoka Y, Webster RG (1989) Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology 169:408–417
Dykhuizen DE, Green L (1991) Recombination in Escherichia coli and the definition of biological species. J Bacteriol 173:7257–7268
Falk BW, Bruening G (1994) Will transgenic crops generate new viruses and new diseases? Science 263:1395–1396
Felsenstein J (1989) PHYLIP-phylogeny inference package. Cladistics 5:164–166
Franck A, Guilley H, Jonard G, Richards K, Hirth L (1980) Nucleotide sequence of cauliflower mosaic virus DNA. Cell 21:285–294
Fütterer J, Gordon K, Bonneville JM, Sanfaçon H, Pisan B, Penswick J, Hohn T (1988) The leading sequence of caulimovirus large RNA can be folded into a large stem-loop structure. Nucleic Acids Res 16:8377–8390
Gal S, Pisan B, Hohn T, Grimsley N, Hohn B (1992) Agroinfection of transgenic plants leads to viable cauliflower mosaic virus by intermolecular recombination. Virology 187:525–533
Gojobori T, Moriyama EN, Kimura M (1990) Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci U S A 87: 10015–10018
Gracia O, Shepherd RJ (1985) Cauliflower mosaic virus in the nucleus of Nicotiana. Virology 146:141–145
Grimsley N, Hohn T, Hohn B (1986) Recombination in a plant virus: template-switching in cauliflower mosaic virus. EMBO J 5:641–646
Hasegawa A, Verver J, Shimada A, Saito M, Goldbach R, Van Kammen A, Miki K, Kameya-Jwaki M, Hibi T (1989) The complete sequence of soybean chlorotic mottle virus DNA and the identification of a novel promoter. Nucleic Acids Res 17:9993–10013
Holland JJ, De La Torre JC, Steinhauer DA (1992) RNA virus populations as quasispecies. Curr Top Microbiol Immunol 176:1–20
Howarth AJ, Gardner RC, Messing J, Shepherd RJ (1981) Nucleotide sequence of naturally occurring deletion mutants of cauliflower mosaic virus. Virology 112:678–685
Howarth AJ, Vandemark GJ (1989) Phylogeny of geminiviruses. J Gen Virol 70:2717–2727
Howell SH, Walker LL, Walden RM (1981) Rescue of in vitro generated mutants of cloned cauliflower mosaic virus genome in infected plants. Nature 293:483–486
Hull R (1980) Structure of the cauliflower mosaic virus genome III. Restriction endonuclease mapping of thirty-three isolates. Virology 100:76–90
Hull R, Sadler J, Longstaff M (1986) The sequence of carnation etched ring virus DNA: comparison with cauliflower mosaic virus and retroviruses. EMBO J 5:3083–3090
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Li W-H, Tanimura M, Sharp PM (1988) Rates and dates of divergence between AIDS virus nucleotide sequences. Mol Biol Evol 5:313–330
Lung MCY, Pirone TP (1972) Datura stramonium, a local lesion host for certain isolates of cauliflower mosaic virus. Phytopathology 62:1473–1474
Mason WS, Taylor JM, Hull R (1987) Retroid virus genome replication. Adv Virus Res 32:35–96
Matthews R (1991) Plant virology, 3rd ed. Academic Press, New York
Melcher U, Choe IS, Lebeurier G, Richards K, Essenberg RC (1986) Selective allele loss and interference between cauliflower mosaic virus DNAs. Mol Gen Genet 203:230–236
Melcher U (1989) Symptoms of cauliflower mosaic virus infection in Arabidopsis thaliana and turnip. Bot Gazette 150:139–147
Melcher U (1990) Similarities between putative transport proteins of plant viruses. J Gen Virol 71:1009–1018
Melcher U, Lartey RT, Pennington RE (1992) Isolate-specific synergy in symptom production between a caulimo- and a tobamovirus. Phytopathology 82:1173–1174
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Pennington RE, Melcher U (1993) In planta deletion of DNA inserts from the large intergenic region of cauliflower mosaic virus DNA. Virology 192:188–196
Penny D, Hendy MD, Steel MA (1991) Testing the theory of descent. In: Miyamoto MM, Cracraft J (eds) Phylogenetic analysis of DNA sequences. Oxford University Press, New York, pp 155–183
Riederer MA, Grimsley NH, Hohn B, Jiricny J (1992) The mode of cauliflower mosaic virus propagation in the plant allows rapid amplification of viable mutant strains. J Gen Virol 73:1449–1456
Rongxiang F, Xiacjun W, Ming B, Yingchuan T, Faxing C, Kequiang M (1985) Complete nucleotide sequence of cauliflower mosaic virus (Xinjing isolate) genomic DNA. Chin J Virol 1:247–256
Sanger M, Daubert S, Goodman RM (1991) The regions of sequence variation in caulimovirus gene VI. Virology 182:830–834
Sawyer S (1989) Statistical tests for detecting gene conversion. Mol Biol Evol 6:526–538
Schoelz J, Shepherd RJ, Daubert S (1986) Region VI of cauliflower mosaic virus encodes a host range determinant. Mol Cell Biol 6: 2632–2637
Schoelz JE, Shepherd RJ (1988) Host range control of cauliflower mosaic virus. Virology 162:30–37
Shepherd RJ (1989) Biochemistry of DNA plant viruses. In: Marcus A (ed) The biochemistry of plants. Academic Press, New York, pp 563–616
Shepherd RJ, Bruening GE, Wakeman RJ (1970) Double-stranded DNA from cauliflower mosaic virus. Virology 41:339–347
Sokal RR, Rohlf FJ (1981) Taxonomic congruence in the Leptodomorpha reexamined. Syst Zool 30:309–325
Steinhauer DA, Holland JJ (1987) Rapid evolution of RNA viruses. Annu Rev Microbiol 41:409–433
Stenger DC, Mullin RH, Morris TJ (1988) Isolation, molecular cloning, and detection of strawberry vein banding virus DNA. Phytopathology 78:154–159
Stratford R, Covey SN (1989) Segregation of cauliflower mosaic virus symptom genetic determinants. Virology 172:451–459
Vaden VR, Melcher U (1990) Recombination sites in cauliflower mosaic virus DNAs: implications for mechanisms of recombination. Virology 177:717–726
Walden RM, Howell SH (1983) Uncut recombinant plasmids bearing nested cauliflower mosaic virus genomes infect plants by intragenomic recombination. Plant Mol Biol 2:27–31
Wintermantel WM, Schoelz JE (1992) Cauliflower mosaic virus is capable of recombination with transgenic Nicotiana bigelovii that contain CaMV coding sequences. Phytopathology 82:1110
Woolston CJ, Covey SN, Penswick JR, Davies JW (1983) Aphid transmission and a polypeptide are specified by a defined region of the cauliflower mosaic virus genome. Gene 23:15–23
Zhang XS, Melcher U (1989) Competition between isolates and variants of cauliflower mosaic virus in infected turnip plants. J Gen Virol 70:3427–3437
Author information
Authors and Affiliations
Additional information
Correspondence to: U. Melcher
Rights and permissions
About this article
Cite this article
Chenault, K., Melcher, U. Phylogenetic relationships reveal recombination among isolates of cauliflower mosaic virus. J Mol Evol 39, 496–505 (1994). https://doi.org/10.1007/BF00173419
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00173419