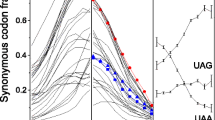
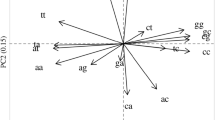
Abstract
We analyzed occurrences of bases in 20,352 introns, exons of 25,574 protein-coding genes, and among the three codon positions in the protein-coding sequences. The nucleotide sequences originated from the whole spectrum of organisms from bacteria to primates. The analysis revealed the following: (1) In most exons, adenine dominates over thymine. In other words, adenine and thymine are distributed in an asymmetric way between the exon and the complementary strand, and the coding sequence is mostly located in the adenine-rich strand. (2) Thymine dominates over adenine not only in the strand complementary to the exon but also in introns. (3) A general bias is further revealed in the distribution of adenine and thymine among the three codon positions in the exons, where adenine dominates over thymine in the second and mainly the first codon position while the reverse holds in the third codon position. The product (A1/T1) × (A2/T2) × (T3/A3) is smaller than one in only a few analyzed genes.
Similar content being viewed by others
References
Aota S, Ikemura T (1986) Diversity in G + C content at the third codon position in vertebrate genes and its cause. Nucleic Acids Res 14: 6345–6355
Bernardi G, Mouchiroud D, Gautier C (1993) Silent substitutions in mammalian genomes and their evolutionary implications. J Mol Evol 37:583–589
Curran JF, Gross BL (1994) Evidence that GHN phase bias does not constitute a framing code. J Mol Biol 235:389–395
Dutton MJ (1985) Genetic code redundancy and the evolutionary stability of protein secondary structure. J Theor Biol 116:343–348
Fickett JW, Torney CT, Wolf DR (1992) Base compositional structure of genomes. Genomics 13:1056–1064
Fickett JW, Tung CS (1992) Assessment of protein coding measures. Nucleic Acids Res 20:6441–6450
Grantham R, Perrin P, Mouchiroud D (1986). Patterns in codon usage of different kinds of species. Oxford Surv Evol Biol 3:48–81
Ikehara K, Okazawa E (1993) Unusually biased nucleotide sequences on sense strands of Flavobacterium sp. genes produce nonstop frames on the corresponding antisense strands. Nucleic Acids Res 21:2193–2199
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Kano A, Ohama T, Abe R, Osawa S (1993) Unassigned or nonsense codons in Micrococcus luteus. J Mol Biol 230:51–56
Kypr J (1986) A part of codon bias in genes protects protein spatial structures from destabilization by random single point mutations. Biochem Biophys Res Commun 139:1094–1097
Kypr J (1990) Possible reason for the preferential insertion of adenine opposite abasic lesions in DNA. J Theor Biol 135:125–126
Kypr J, Mrázek J (1987a) Occurrence of nucleotide triplets in genes and secondary structure of the coded proteins. Int J Biol Macromol 9:49–53
Kypr J, Mrázek J (1987b) Unusual codon usage of HIV. Nature 327: 20
Kypr J, Mrázek J, Reich J (1989) Nucleotide composition bias and CpG dinucleotide content in the genomes of HIV and HTLV 1/2. Biochim Biophys Acta 1009:280–282
Lacey JC, Hall LM, Mullins DW (1985) Rationalization of some genetic anticodonic assignments. Orig Life 16:69–79
Lagunez-Otero J, Trifonov EN (1992) mRNA periodical infrastructure complementary to the proofreading site in the ribosome. J Biomol Struct Dyn 10:455–464
Leskiw BW, Bibb MJ, Chater KF (1991) The use of a rare codon specifically during development? Mol Microbiol 5:2861–2867
Mrázek J, Kypr J (1992) Nucleotide composition of genes and hydrophobicity of the encoded proteins. FEBS Lett 305:163–165
Morijama EN, Gojobori T (1992) Rates of synonymous substitution and base composition of nuclear genes in Drosophila. Genetics 130:855–864
Ohkubo S, Muto A, Kawauchi Y, Yamao F, Osawa S (1987) The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet 210:314–322
Randall SK, Eritja R, Kaplan BE, Petruska J, Goodman MF (1987) Nucleotide insertion kinetics opposite a basic lesions in DNA. J Biol Chem 262:6864–6870
Rice CM, Fuchs R, Higgins DG, Stoehr PJ, Cameron GN (1993) The EMBL Data Library. Nucleic Acids Res 21:2967–2971
Sagher D, Strauss B (1983) Insertion of nucleotides opposite apurinic/ apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry 22:4518–4526
Sharp PM, Li W-H (1987) The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential application. Nucleic Acids Res 15:1281–1295
Shepherd JCW (1981) Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc Natl Acad Sci USA 78:1596–1600
Soto MA, Sepúlveda A, Tohá J (1985) Conservation of the secondary structure of protein during evolution and the role of the genetic code. Orig Life 16:157–164
Stephens RM, Schneider TD (1992) Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J Mol Biol 228:1124–1136
Taylor FJR, Coates D (1989) The code within codons. Biosystems 22:177–187
Trifonov EN (1987) Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16S rRNA nucleotide sequences. J Mol Biol 194:643–652
Veaute X, Fuchs RPP (1993) Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science 261:598–600
Volkenstein MV (1966) The genetic coding of the protein structure. Biochim Biophys Acta 119:421–424
Wada K, Wada Y, Doi H, Ishibashi F, Gojobori T, Ikemura T (1991) Codon usage tabulated from the GenBank genetic data. Nucleic Acids Res 19:1981–1986
Weber AL, Lacey JC Jr (1978) Genetic code correlations: amino acids and their anticodon nucleotides. J Mol Evol 11:199–211
Weber JL (1987) Analysis of sequences from the extremely A + T-rich genome of Plasmodium falciparum. Gene 52:103–109
Woese CR, Dugre DH, Saxinger WC, Dugre SA (1966) The molecular basis for the genetic code. Proc Natl Acad Sci USA 55:966–974
Yomo T, Urabe I, Okada H (1992) No stop codons in the antisense strands of the genes for nylon oligomer degradation. Proc Nall Acad Sci USA 89:3780–3784
Author information
Authors and Affiliations
Additional information
Correspondence to: J. Kypr
Rights and permissions
About this article
Cite this article
Mrázek, J., Kypr, J. Biased distribution of adenine and thymine in gene nucleotide sequences. J Mol Evol 39, 439–447 (1994). https://doi.org/10.1007/BF00173412
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00173412