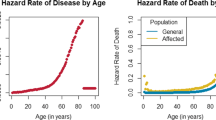
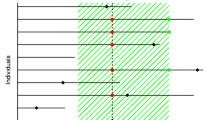
Abstract
The field of genetic epidemiology is growing rapidly with the realization that many important diseases are influenced by both genetic and environmental factors. For this reason, pedigree data are becoming increasingly valuable as a means of studying patterns of disease occurrence. Analysis of pedigree data is complicated by the lack of independence among family members and by the non-random sampling schemes used to ascertain families. An additional complicating factor is the variability in age at disease onset from one person to another. In developing statistical methods for analysing pedigree data, analytic results are often intractable, making simulation studies imperative for assessing the performance of proposed methods and estimators. In this paper, an algorithm is presented for simulating disease data in pedigrees, incorporating variable age at onset and genetic and environmental effects. Computational formulas are developed in the context of a proportional hazards model and assuming single ascertainment of families, but the methods can be easily generalized to alternative models. The algorithm is computationally efficient, making multi-dataset simulation studies feasible. Numerical examples are provided to demonstrate the methods.
Similar content being viewed by others
References
Cannings, C. and Thompson, E. A. (1977) Ascertainment in the sequential sampling of pedigrees. Clinical Genetics, 12, 208–212.
Clayton, D. G. (1991) A Monte Carlo method for Bayesian inference in frailty models. Biometrics, 47, 467–486.
Cox, D. R. (1972) Regression models and life tables (with discussion). Journal of the Royal Statistical Society Series B, 34, 187–220.
Elston, R. and Bonney, G. (1986) Sampling via probands in the analysis of family studies. Proceedings of the 13th International Biometric Conference, 1–14.
Geman, S. and Geman, D. (1984). Stocastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Transactions on Pattern Analysis and Machine Intelligence, 6, 721–741.
Gilks, W. R. (1992) Derivative-free adaptive rejection sampling for Gibbs sampling. Bayesian Statistics, 4, 1–9.
Kalbfleisch, J. D. and Prentice, R. L. (1980) The statistical analysis of failure time data. Wiley, New York.
Pericak-Vance, M. A., Bebout, J. L., Gaskell, P. C. Jr, Yamaoka, L. H., Hung, W.-Y., Alberts, M. J., Walker, A. P., et al. (1991) Linkage studies in familial Alzheimer's disease: evidence for chromosome 19 linkage. American Journal of Human Genetics, 48, 1034–1050.
Rao, C. R. (1973) Linear statistical inference and its applications. Wiley, New York.
Thompson, E. A. (1986) Pedigree analysis in human genetics. Johns Hopkins University Press, Baltimore, MD.
Wichmann, B. A. and Hill, I. D. (1982) An efficient and portable pseudo-random number generator. Applied Statistics, 31, 188–190.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gauderman, W.J. A method for simulating familial disease data with variable age at onset and genetic and environmental effects. Stat Comput 5, 237–243 (1995). https://doi.org/10.1007/BF00142665
Issue Date:
DOI: https://doi.org/10.1007/BF00142665