Abstract
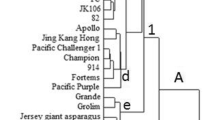
Genetic diversity of eighteen accessions of Ae. geniculata (2n=28; UUMM) was assessed using the random amplified polymorphic DNA (RAPD) technique. We optimized RAPD conditions including the template DNA, the concentration of AmpliTaq DNA polymerase Stoffel fragment, and MgCl2 concentration for revealing polymorphisms. Thirty-eight decamer oligonucleotides were individually used as primers under optimized conditions. Seventeen of these primers produced polymorphic RAPDs among the 18 accessions of Ae. geniculata. Polymorphisms were recorded by noting presence or absence of an amplification product from the total genomic DNA. Comparisons of unique and shared amplification products of each pair of accessions were used to generate genetic similarity coefficients (GSCs). These GSCs were used to construct a phenogram using an unweighted pair-group method with arithmetical averages (UPGMA). The phenogram shows that RAPD data is useful in the measurement of genetic variation or similarity within a species. It also indicates that we can select eight or nine accessions of the eighteen accessions to maintain at least 80% genetic variability of the Chinese collection of Ae. geniculata.
Similar content being viewed by others
References
Bailey, K.L., H. Harding & D.R. Knott, 1993. Transfer to bread wheat of resistance to common root rot (Cochliobolus sativus) identified in Triticum timopheevii and Aegilops ovata. Can. J. Plant Pathol. 15: 211–219.
Devos, K.M. & M.D. Gale, 1992. The use of random amplified polymorphic DNA markers in wheat. Theor. Appl. Genet. 84: 567–572.
Fernandez-Caivin, B. & J. Greliana, 1992. Relationship between pairing frequencies and genome affinity estimations in Aegilops ovata x Triticum aestivum hybrids plants. Heredity 68: 165–172.
Friebe, B. & M. Haun, 1989. C-banding and powdery resistance of Triticum ovatum and four T. aestivum-T. ovatum chromosome addition lines. Theor. Appl. Genet. 78: 417–424.
Gill, B.S., H.C. Sharma, W.J. Raupp, L.E. Browder, J.H. Hatchett, T.L. Harvey, J.G. Moseman & J.G. Waines, 1985. Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, Hessian fly and greenbug. Plant Dis. 69: 314–316.
Joshi, C.P. & H.T. Nguyen, 1993. Application of the random amplified polymorphic DNA technique for the detection of polymorphism among wild and cultivated tetraploid wheats. Genome 36: 602–609.
Kinoshita, T., 1988. Utilization of nucleo-cytoplasmic hybrids with Aegilops ovata cytplasm in wheat breeding. In: T.E. Miller & R.M.D, Koebner (Eds), Proc. 7th Int. Wheat Genet. Symp., Cambridge, U.K. pp. 1133–1138.
Kinoshita, T. & H. Kihara, 1983. Nucleo-cytoplasmic heterosis in common wheat having Aegilops ovata cytoplasm. In: S. Sakamoto, (Ed.), Proc. 6th Int. Wheat Genet. Symp., Kyoto. pp. 507–512.
Kressovich, S., J.G.K. Williams, J.R. McFerson, E.J. Routman & B.A. Schael, 1992. Characterization of genetic identities and relationships of Brassica oleracea L. via a random amplified polymorphic DNA assay. Theor. Appl. Genet. 85: 190–196.
Muralidharan, K. & E.K. Wakeland, 1993. Concentration of primer and template qualitatively affects products in random-amplified polymorphic DNA PCR. BioTechniques, 14: 362–364.
Park, Y.H. & R.J. Kohel, 1994. Effect of concentration of MgCl2 on random-amplified DNA polymorphism. BioTechniques. 16: 652–653.
Rogers, S.C. & A.J. Bendich, 1988. Extraction of DNA from plant tissue. In: Plant Molecular Biology Manual, A6: 1–10. Kluwer Academic Publishers. Dordrecht.
Rohlf, F.J., 1993. NTSYS-pc: Numerical taxonomy and multivariate analysis system. Applied Biostatistics Inc., New York.
Skroch, P., J. Tivang & J. Nienhuis, 1992. Analysis of genetic relationships using RAPD marker data. In: Applications of RAPD technology to plant breeding. Joint Plant Breeding Symposia Series, ASA, CSSA, & SSSA, Maddison, WI. pp. 26–30.
Vierling, R.A. & H.T. Nguyen, 1992. Use of RAPD markers to determine the genetic diversity of diploid wheat genotypes. Theor. Appl. Genet. 84: 835–838.
Wei, J.Z. & R.R-C. Wang, 1994. Genetic variability in Russian wild rye (Psathyrostachys juncea) assessed by RAPD. Agron. Abstr. ASA, CSSA, SSSA, Maddison, WI. p. 222.
Williams, J.G.K., A.R. Kubelic, K.J. Livak, J.A. Rafalski & S.V. Tingey, 1990. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Res. 18: 6531–6535.
Yang, T.Z. & S.S. Maan, 1985. Possibility of utilizing Ae. ovata cytoplasm to combine two kinds of heterosis in wheat. Acta Agri. Univ. Pekinensis 11: 199–207.
Zhang, X.Y., Y.S. Dong & R. R-C Wang, 1994. Geographic distribution of alleles for esterase-5, gliadin, α- and β-amylase in Triticum tauchii. Proc. 2nd Int. Triticeae Symp., Logan, Utah (in press).
Author information
Authors and Affiliations
Additional information
Address for correspondence: Dr X-Y. Zhang, USDA-ARS-FRRL, Utah State University, Logan, UT 84322-6300, who is visiting the USA under an agreement between USDA-ARS and CMA-CAAS on germplasm resources.
Rights and permissions
About this article
Cite this article
Zhang, X.Y., Wang, R.R.C. & Dong, Y.S. RAPD polymorphisms in Aegilops geniculata Roth (Ae. ovata auct. non L.). Genet Resour Crop Evol 43, 429–433 (1996). https://doi.org/10.1007/BF00123733
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00123733