Summary
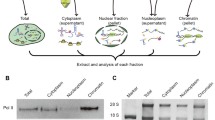
A simple and rapid procedure has been developed for the isolation of chromatin from plant leaves. The molecular weight of the DNA extracted from these chromatin preparations is comparable to that of DNA isolated by a conventional purification procedure (CTAB-CsCl-method). These results suggest that almost no degradation occurs during the isolation procedure. The effect of DNase I on three different groups of genes was studied; one of them, encoding the NADPH-protochlorophyllide oxidoreductase (PCR), represents a gene which is actively transcribed in etiolated leaf tissue. The other genes examined encode the hordein seed storage protein and 26S ribosomal RNA. The hordein genes are known to be inactive in leaves.
The hordein and rDNA genes were found to be resistant to low levels of DNase I, while the gene for the PCR was highly sensitive to DNase I. During the course of digestion of the PCR gene, discrete cleavage products are generated. These indicate the presence of DNase I hypersensitive sites in the vicinity of the PCR gene in etiolated leaves. As a control ‘naked’ DNA has been digested with DNase I. No differences in sensitivity between the PCR gene and the hordein genes can be detected.
Similar content being viewed by others
References
Apel K: The protochlorophyllide holochrome of barley (Hordeum vulgare L.) Phytochrome induced decrease of translatable mRNA coding for the NADPH-protochlorophyllide oxidoreductase. Eur J Biochem 120:89–93, (1981).
Apel K, Gollmer I, Batschauer A: The light-dependent control of chloroplast development in barley (Hordeum vulgare L.). J Cell Biochem 23:181–189, 1983.
Appels R, Gerlach WL, Dennis ES, Swift H, Peacock WJ: Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 78:293–311, 1980.
Chen Y-M, Lin C-Y, Chang H, Guilfoyle TJ, Key JL: Isolation and properties of nuclei from control and auxin-treated soybean hypocotyl. Plant Physiol 56:78–82, 1975.
Dretzen G, Bellard M, Sassone-Corsi P, Chambon P: A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem 112:295–298, 1981.
Elgin SCR: DNase I-hypersensitive sites in chromatin. Cell 27:413–415, 1981.
Elgin SCR: Anatomy of hypersensitive sites. Nature 309:213–214, 1984.
Ferl RJ: Modulation of chromatin structure in the regulation of the maize Adh 1 gene. Mol Gen Genet 200:207–210, 1985.
Flavell RB, O'Dell M, Smith DB, Thompson WF: Chromosome architecture: the distribution of recombination sites, the structure of ribosomal DNA loci and the multiplicity of sequences containing inverted repeats. In: Molecular Form and Function of the Plant Genome (van Vloten-Doting L, Groot GSP, Hall TC, eds.) NATO ASI series. Series A. Life sciences; Vol. 83; pp. 1–14, 1985, Plenum Press, New York, London.
Forde BG, Kreis M, Bahramian MB, Matthews IA, Miflin BJ, Thompson RD, Bartels D, Flavell RB: Molecular cloning and analysis of cDNA sequences derived from polyA+ RNA from barley endosperm: identification of B hordein related clones. Nucleic Acids Res 9:6689–6707, 1981.
Fritton HP, Igo-Kemenes T, Navock J, Strech-Jurk U, Theisen M, Sippel AE: Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme. Nature 311:163–165, 1984.
Garel A, Axel R: Selective digestion of transcriptionally active ovalbumin from oviduct nuclei. Proc Natl Acad Sci USA 73:3966–3970, 1976.
Hopp HE, Rasmussen SK. Brandt A: Organization and transcription of B 1 hordein genes in high lysine mutants of barley. Carlsberg Res Comm 48:201–216, 1983.
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning, Cold Spring Harbor, Laboratory, New York, 1982.
Miflin BJ, Rahman S, Kreis N, Forde BG, Blanco L, Shewry PR: The hordein of barley: Developmentally and nutritionally regulated multigene families of storage proteins. In: Ciferri O, Dure L III (eds) Structure and Function of Plant Genomes, NATO ASI series. Series A, Life sciences; Vol. 63: pp. 85–92, 1983. Plenum Press, New York, London.
Mösinger E, Batschauer A, Schäfer E, Apel K: Phytochrome control of in vitro transcription of specific genes in isolated nuclei from barley (Hordeum vulgare) Eur J Biochem 147:137–142, 1985.
Murray MG, Kennard WC: Altered chromatin conformation of the higher plant gene phaseolin. Biochemistry 23:4225–4232, 1984.
Rasmussen SK, Hopp HE, Brandt A: Nucleotide sequences of cDNA clones for B 1 hordein polypeptides. Carlsberg Res Comm 48:187–199, 1983.
Reeves R: Transcriptionally active chromatin. Biochim et Biophys Acta 782:343–393, 1984.
Samal B, Worcel A, Louis C, Schedl P: Chromatin structure of the histone genes of D. melanogaster. Cell 23:401–409, 1981.
Santel HJ, Apel K: The protochlorophyllide holochrome of barley (Hordeum vulgare L.). The effect of light on the NADPH: protochlorophyllide oxidoreductase. Eur J Biochem 120:95–103, 1981.
Spiker S, Murray MG, Thompson WF: DNase I sensitivity of transcriptionally active genes in intact nuclei and isolated chromatin of plants. Proc Natl Acad Sci USA 80:815–819, 1983.
Spiker S: Chromatin structure and gene regulation in higher plants. Adv Genet 22:145–208, 1984.
Spiker S: Plant chromatin structure. Ann Rev Plant Physiol 36:235–253, 1985.
Stalder J, Groudine M, Dodgson JB, Engel JD, Weintraub H: Hb switching in chickens. Cell 19:973–980, 1980.
Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H: Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNase I. Cell 20:451–460, 1980b.
Taylor B, Powell A: Isolation of plant DNA and RNA BRL Focus 4:4–6, 1982.
Weintraub H, Groudine M: Chromosomal subunits in active genes have an altered conformation. Science 193:848–856, 1976.
Weisbrod S: Active chromatin. Nature 297:289–295 1982.
Wieslander L: A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem 98:305–309, 1979.
Willmitzer L, Wagner KG: The isolation of nuclei from tissuecultured plant cells. Exp Cell Res 135:69–77, 1981.
Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SCR: The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell 16:797–806, 1979.
Wu C: The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286: 854–860, 1980.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Steinmüller, K., Apel, K. A simple and efficient procedure for isolating plant chromatin which is suitable for studies of DNase I-sensitive domains and hypersensitive sites. Plant Mol Biol 7, 87–94 (1986). https://doi.org/10.1007/BF00040135
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00040135