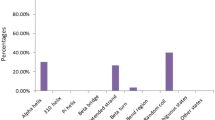
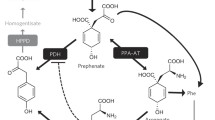
Abstract
Seeds of Pisum sativum contain a biotinyl polypeptide called SBP65 that behaves as a putative sink for the free vitamin, representing more than 90% of the total protein-bound biotin in mature seeds. A cDNA encoding SBP65 was cloned and sequenced. The deduced primary structure of the protein was confirmed by protein sequencing. Peptide sequencing also indicated binding of the biotin to lysine 103. The biotinylation domain of SBP65 differs markedly from that of presently known biotin enzymes. Molecular analysis of the protein sequence reveals an extremely hydrophilic protein containing several repeated motifs. These properties, as well as the temporal and spatial patterns of expression of this protein, suggest that SBP65 belongs to the LEA (late embryogenesis-abundant) group of proteins.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 215: 403–410 (1990).
Al-Feel W, Chirala SS, Wakil SJ: Cloning of the yeast FAS3 gene and primary structure of yeast acetyl CoA carboxylase. Proc Natl Acad Sci USA 89: 4534–4538 (1992).
Ashton AR, Jenkins CL, Whitfeld PR: Patent WO 93/11243 (1993).
Best EA, Knauf VC: Organization and nucleotide sequence of the genes encoding the biotin carboxyl carrier protein and biotin carboxylase protein of Pseudomonas aeruginosa acetyl-CoA carboxylase. J Bact 175: 6881–6889 (1993).
Brocklehurst SM, Perham RN: Prediction of the three-dimensional structure of the biotinylated domain from pyruvate carboxylase and of the lipoylated H-protein from the pea leaf glycine cleavage system: a new automated method for the prediction of protein tertiary structure. Protein Sci 2: 626–639 (1993).
Browner MF, Taroni F, Sztul E, Rosenberg LE: Sequence analysis, biogenesis and mitochondrial import of the alpha-subunit of rat liver propionyl-CoA carboxylase. J Biol Chem 264: 12680–12685 (1989).
Chandler CS, Ballard FJ: Regulation of the breakdown rates of biotin-containing proteins in Swiss 3T3-L1 cells. Biochem J 251: 749–755 (1988).
Craft DV, Goss NH, Chandramouli N, Wood HG: Purification of biotinidase from human plasma and its activity on biotinyl peptides. Biochemistry 24: 2471–2476 (1985).
Dakshinamurti K, Chauhan J: Vitamins and Hormones 45: 337–384 (1989).
Daniel JHAS, Kong I-S, Park C-K, Tac H-J, Kim K-H: Cloning of human acetyl-CoA carboxylase cDNA. Eur J Biochem 219: 297–306 (1994).
Dure LIII, Crouch M, Harada J, Ho T-HD, Mundy J, Quatrano R, Thomas T, Sung ZR: Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol 12: 475–486 (1989).
Duval M, Job C, Alban C, Douce R, Job D: Developmental patterns of free and protein-bound biotin during maturation and germination of seeds of Pisum sativum: characterization of a novel seed-specific biotinylated protein. Biochem J 299: 141–150 (1994).
Duval M, Job C, Alban C, Sparace S, Douce R, Job D: Synthesis and degradation of a novel biotinyl protein in developing and germinating pea seeds. C R Acad Sci (Paris) Sér III 316: 1463–1470 (1993).
Fall RR: Analysis of microbial biotin proteins. Meth Enzymol 67: 390–398 (1979).
Galau GA, Jakobsen KJ, Hughes DW: The controls of late dicot embryogenesis and early germination. Physiol Plant 81: 280–288 (1991).
Glazer AN, Delange RJ, Sigman DS: Modification of protein side-chains: Group-specific reagents. In: Work TS, Work E (eds) Chemical Modifications of Proteins: Selected Methods and Analytical Procedures, pp. 68–120. North-Holland Publishing Company, Amsterdam (1975).
Gornicki P, Scappino LA, Haselkorn R: Genes for two subunits of acetyl coenzyme A carboxylase of Anabaena sp. strain PCC 7120: biotin carboxylase and biotin carboxyl carrier protein. J Bact 175: 5268–5272 (1993).
Guchaiiait RB, Polakis EF, Dimroth P, Stoll E, Moss J, Lane MD: Acetyl coenzyme A carboxylase system of Escherichia coli. Purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J Biol Chem 249: 6633–6645 (1974).
Harwood JL: Fatty acid metabolism. Annu Rev Plant Physiol Plant Mol Biol 39: 101–138 (1988).
Harwood JL: The site of action of some selective graminaceous herbicides is identified as acetyl-CoA carboxylase. Trends Biochem Sci 13: 330–331 (1988).
Knowles JR: The mechanism of biotin-dependent enzymes. Annu Rev Biochem 58: 195–221 (1989).
Kondo H, Uno S, Komizo Y, Sunamoto J: Importance of methionine residues in the enzymatic carboxylation of biotin-containing peptides representing the local biotinyl site of E. coli acetyl-CoA carboxylase. Int J Peptide Protein Res 23: 559–564 (1984).
Kyte J, Doolittle RF: A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 (1982).
Li S-J, CronanJr JE: The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267: 855–863 (1992).
Lopez-Casillas F, Bai D-H, Luo X, Kong IS, Hermodson MA, Kim K-H: Structure of the coding sequence and primary amino acid sequence of rat acetyl coenzyme A carboxylase. Proc Natl Acad Sci USA 85: 5784–5788 (1988).
Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA: Selection of AUG initiation codons differs in plants and animals. EMBO J 6: 43–48 (1987).
Moss J, Lane MD: The biotin-dependent enzymes. Adv Enzymol 35: 321–442 (1971).
Newman W, Beall LD, Randhawa ZI: Biotinylation of peptide hormones: Structural analysis and application to flow cytometry. Meth Enzymol 184: 275–285 (1990).
Parker WB, Marshall LC, Burton JD, Somers DA, Wyse DL, Gronwald JW, Gengenbach BG: Dominant mutations causing alterations in acetyl-coenzyme A carboxylase confer tolerance to cyclohexanedione and aryloxyphenoxypropionate herbicides in maize. Proc Natl Acad Sci USA 87: 7175–7179 (1990).
Puupponen-Pimiä R, Saloheimo M, Vasara T, Ra R, Gaugecz J: Characterization of a birch (Beta pendula Roth.) embryogenic gene, BP8. Plant Mol Biol 23: 423–428 (1993).
Rendina AR, Felts JM: Cyclohexanedione herbicides are selective and potent inhibitors of acetyl-CoA carboxylase from grasses. Plant Physiol 86: 983–986 (1988).
Robinson BH, Oei J, Saunders M, Gravel R: 3H-Biotinlabeled proteins in cultured human skin fibroplasts from patients with pyruvate carboxylase deficiency. J Biol Chem 258: 6660–4 (1983).
Roessler PG, Ohlrogge JB: Cloning and characterization of the gene that encodes acetyl-coenzyme A carboxylase in the alga Cyclotella cryptica. J Biol Chem 268: 19254–19259 (1993).
Samols D, Thornton CG, Murtif VL, Kumar GK, Haase FC, Wood HG: Evolutionary conservation among biotin enzymes. J Biol Chem 263: 6461–6464 (1988).
Schatz PJ: Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Bio/technology 11: 1138–1143 (1993).
Scheiner J, De Ritter E: Biotin content of feedstuffs. J Agric Food Chem 23: 1157–1162 (1975).
Schwarz E, Oesterhelt D, Reinke H, Beyreuther K, Dimroth P: The sodium ion translocating oxalacetate decarboxylase of Klebsiella pneumoniae. Sequence of the biotin-containing α-subunit and relationship to other biotin-containing enzymes. J Biol Chem 263: 9640–9645 (1988).
Secor J, Cséke C: Inhibition of acetyl-CoA carboxylase activity by haloxyfop and traloxydim. Plant Physiol 86: 10–12 (1988).
Shenoy BC, Paranjape S, Murtif VI, Kumar GK, Samols D, Wood HG: Effect of mutations at Met-88 and Met-90 on the biotination of Lys-89 of the apo 1.3S subunit of transcarboxylase. FASEB J 2: 2505–2511 (1988).
Shriver BJ, Roman-Shriver C, Allred JB: Depletion and repletion of biotinyl enzymes in liver of biotin-deficient rats: Evidence of a biotin storage system. J Nutr 123: 1140–1149 (1993).
White HBIII, Whitehead CC: Role of avidin and other biotin-binding proteins in the deposition and distribution of biotin in chicken eggs: discovery of a new biotin-binding protein. Biochem J 241: 677–684 (1987).
Wang D, Waye MMY, Taricani M, Buckingham K, Sandham HJ: Biotin-containing protein as a cause of false positive clones in gene probing with streptavidin/biotin. Bio/Techniques 14: 209–212 (1993).
Woehlke G, Wifling K, Dimroth P: Sequence of the sodium ion pump oxalacetate decarboxylase from Salmonella typhimurium. J Biol Chem 267: 22798–22803 (1992).
Wurtele ES, Nikolau BJ: Plants contain multiple biotin enzymes. Discovery of 3-methylcrotonyl-CoA carboxylase, propionyl-CoA carboxylase and pyruvate carboxylase in the plant kingdom. Arch Biochem Biophys 278: 179–186 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Duval, M., DeRose, R.T., Job, C. et al. The major biotinyl protein from Pisum sativum seeds covalently binds biotin at a novel site. Plant Mol Biol 26, 265–273 (1994). https://doi.org/10.1007/BF00039537
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00039537