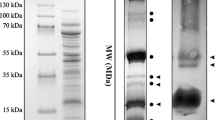
Abstract
A protein fraction which lacks DNA-binding activity itself, but confers enhanced protein-DNA complex formation to E. coli core RNA polymerase, was obtained from mustard chloroplasts by heparin Sepharose chromatography. Gel retardation and competition assays as well as DNase I footprinting experiments with a chloroplast DNA fragment containing the psbA promoter indicate that this reflects sequence-specific binding. Transcription of the psbA template by E. coli core enzyme in the presence of the chloroplast fraction results in enhanced formation of transcripts of the size expected for correct initiation at the in vivo start site. We conclude that the chloroplast fraction reveals sigma-like activity with E. coli RNA polymerase and thus might contain factor(s) of equivalent function in chloroplast transcription.
Similar content being viewed by others
References
Berg D, Barrett K, Chamberlin M: Purification of two forms of Escherichia coli RNA polymerase and of sigma component. In: Grossman L, Moldave K (eds) Methods in Enzymology, Vol. 65. Academic Press, New York (1971) pp 499–525.
Bickle TA, Pirrotta V, Imber R: A simple general procedure for purifying restriction endonucleases. Nucleic Acids Res 4: 2561–2572 (1977).
Boyer SK, Mullet JE: Characterization of P. sativum chloroplast psbA transcripts produced in vivo, in vitro and in E. coli. Plant Mol Biol 6: 229–243 (1986).
Bradley D, Gatenby AA: Mutational analysis of the maize chloroplast ATPase-β subunit gene promoter: the isolation of promoter mutants in E. coli and their characterization in a chloroplast in vitro transcription system. EMBO J 4: 3641–3648 (1985).
Briat JF, Dron M, Loiseaux S, Mache R: Structure and transcription of the spinach chloroplast rDNA leader region. Nucleic Acids Res 10: 6865–6878 (1982).
Briat JF, Lescure AM, Mache R: Transcription of the chloroplast DNA: a review. Biochimie 68: 981–990 (1986).
Burgess RR, Travers AA, Dunn JJ, Bautz EKF: Factor stimulating transcription by RNA polymerase. Nature 221: 43–46 (1969).
Carthew RW, Chodosh LA, Sharp PA: An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell 43: 439–448 (1985).
Cozens AL, Walker E: Pea chloroplast DNA encodes homologues of Escherichia coli ribosomal subunit S2 and the β′-subunit of RNA polymerase. Biochem J 236: 453–460 (1986).
Duval-Valentin G, Ehrlich R: Interaction between E. coli RNA polymerase and the tetR promoter from pSC101: homologies and differences with other E. coli promoter systems from close contact point studies. Nucleic Acids Res 14: 1967–1983 (1986).
Fried M, Crothers DM: Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res 9: 6505–6525 (1981).
Galas DJ, Schmitz A: DNAase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res 5: 3157–3168 (1978).
Garner MM, Revzin A: A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res 9: 3047–3060 (1981).
Garoff H, Ansorge W: Improvements of DNA sequencing gels. Anal Biochem 115: 450–457 (1981).
Greenberg BM, Narita JO, DeLuca-Flaherty CR, Hallick RB: Properties of chloroplast RNA polymerases. In: Steinback KE, Bonitz S, Arntzen C, Bogorad L (eds) Molecular Biology of the Photosynthetic Apparatus. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1985) pp 303–310.
Gruissem W, Zurawski G: Analysis of promoter regions for the spinach chloroplast rbcL, atpB and psbA genes. EMBO J 4: 3375–3383 (1985).
Heumann H, Metzger W, Niehörster M: Visualization of intermediary transcription states in the complex between Escherichia coli DNA-dependent RNA polymerase and a promoter-carrying DNA fragment using the gel retardation method. Eur J Biochem 158: 575–579 (1986).
Hofer B, Müller D, Köster H: The pathway of E. coli RNA polymerase-promoter complex formation as visualized by footprinting. Nucleic Acids Res 13: 5995–6013 (1985).
Jolly SO, Bogorad L: Preferential transcription of cloned maize chloroplast DNA sequences by maize chloroplast RNA polymerase. Proc Natl Acad Sci USA 77: 822–826 (1980).
Kidd GH, Bogorad L: A facile procedure for purifying maize chloroplast RNA polymerase from whole cell homogenates. Biochim Biophys Acta 609: 14–30 (1980).
Kung SD, Lin CM: Chloroplast promoters from higher plants. Nucleic Acids Res 13: 7543–7549 (1985).
Lerbs S, Bräutigam E, Parthier B: Polypeptides of DNA-dependent RNA polymerase of spinach chloroplasts: characterization by antibody-linked polymerase assay and determination of sites of synthesis. EMBO J 4: 1661–1666 (1985).
Lerbs S, Briat JF, Mache R: Chloroplast RNA polymerase from spinach: purification and DNA-binding proteins. Plant Mol Biol 2: 67–74 (1983).
Link G: DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro system from mustard (Sinapis alba L.). EMBO J 3: 1697–1704 (1984).
Link G, Langridge U: Structure of the chloroplast gene for the precursor of the Mr 32000 photosystem II protein from mustard (Sinapis alba L.). Nucleic Acids Res 12: 945–957 (1984).
Losick R, Chamberlin MJ: RNA Polymerase. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1976).
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning, a Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1982).
Maxam A, Gilbert W: Sequencing end-labeled DNA with base-specific chemical cleavages. In: Grossman L, Moldave K (eds) Methods in Enzymology, Vol. 65. Academic Press, New York (1980) pp 499–525.
Neuhaus H, Link G: The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet 11: 251–257 (1987).
Ohme M, Tanaka M, Chunwongse J, Shinozaki K, Sugiura M: A tobacco chloroplast DNA sequence possibly coding for a polypeptide similar to E. coli RNA polymerase β-subunit. FEBS Lett 200: 87–90 (1986).
Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano K, Umesono L, Shiki Y, Takeuchi M, Chang Z, Hota S, Inokuchi M, Ozeki H: Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322: 572–574 (1986).
Orozco EM, Mullet JE, Chua NH: An in vitro system for accurate transcription initiation of chloroplast protein genes. Nucleic Acids Res 13: 1283–1302 (1985).
Reiss T, Link G: Characterization of transcriptionally active DNA-protein complexes from chloroplasts and etioplasts of mustard (Sinapis alba L.). Eur J Biochem 148: 207–212 (1985).
Sanger F, Coulson AR: The use of thin acrylamide gels for DNA sequencing. FEBS Lett 87: 107–108 (1978).
Schwarz Z, Kössel H, Schwartz E, Bogorad L: A gene coding for tRNAVal is located near 5′ terminus of 16 S rRNA gene in Zea mays chloroplast genome. Proc Natl Acad Sci USA 78: 4748–4752 (1981).
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazada K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M: The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5: 2043–2049 (1986).
Sijben-Müller G, Hallick RB, Alt J, Westhoff P, Herrmann RG: Spinach plastid genes coding for initiation factor IF-1, ribosomal protein S11 and RNA polymerase α-subunit. Nucleic Acids Res 14: 1029–1044 (1986).
Smith HJ, Bogorad L: The polypeptide subunit structure of the DNA-dependent RNA polymerase of Zea mays chloroplasts. Proc Natl Acad Sci USA 71: 4839–4842 (1974).
Straney DC, Crothers DM: Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell 43: 449–459 (1985).
Surzycki SJ, Shellenbarger DL: Purification and characterization of a putative sigma factor from Chlamydomonas reinhardii. Proc Natl Acad Sci USA 73: 3961–3965 (1976).
Tewari KK, Goel A: Solubilization and partial purification of RNA polymerase from pea chloroplasts. Biochemistry 22: 2142–2148 (1983).
Whitfeld PR, Bottomley W: Organization and structure of chloroplast genes. Annu Rev Plant Physiol 34: 279–310 (1983).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bülow, S., Link, G. Sigma-like activity from mustard (Sinapis alba L.) chloroplasts conferring DNA-binding and transcription specificity to E. coli core RNA polymerase. Plant Mol Biol 10, 349–357 (1988). https://doi.org/10.1007/BF00029885
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00029885