Abstract
A structural gene encoding nitrite reductase (NiR) in bean (Phaseolus vulgaris) has been cloned and sequenced. The NiR gene is present as a single copy encoding a protein of 582 amino acids. The bean NiR protein is synthezised as a precursor with an amino-terminal transit peptide (TP) consisting of 18 amino acid residues. The bean NiR transit peptide shows similarity to the TPs of other known plant NiRs.
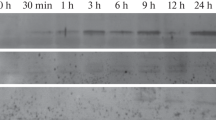
The NiR gene is expressed in trifoliate leaves and in roots of 20-day old bean plants where transcript accumulation is nitrate-inducible. Gene expression occurs in a circadian rhythm and induced by light in leaves of dark-adapted plants.
A particular 100 bp sequence is present in the promoter and in the first intron of the NiR gene. Several copies of this 100 bp sequence are present in the bean genome. Comparisons between the promoter of the bean NiR gene and of two bean nitrate reductase genes (NR1 and NR2) show a limited number of conserved motifs, although the genes are presumed to be co-regulated. Comparisons are also made between the bean NiR promoter and the spinach NiR promoter.
Transformation of tobacco plants with the bean NiR promoter fused to the GUS reporter gene (β-glucuronidase) shows that the bean NiR promoter is nitrate-regulated and that the presence of the 100 bp sequence influences the level of GUS activity. NiR-coding sequences are not required for nitrate regulation but have a quantitative effect on the measured GUS activity.
Similar content being viewed by others
References
Aslam M, Huffaker RC: Role of nitrate and nitrite in the induction of nitrite reductase in leaves of barley seedlings. Plant Physiol 91: 1152–1156 (1989).
Augier V, Guigliarelli B, Asso M, Bertrand P, Froxon C, Giordano G, Chippaux M, Blasco F: Site-directed mutagenesis of conserved cysteine residues within the β-subunit of Escherichia coli nitrate reductase. Physiological, biochemical and EPR characterization of the mutated enzymes. Biochemistry 32: 2013–2023 (1993).
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Strul K: Current Protocols in Molecular Biology. Greene Wiley-Interscience, New York (1987).
Back E, Burkhart W, Moyer M, Privalle L, Rothstein S: Isolation of cDNA clones coding for spinach nitrite reductase: complete sequence and nitrate induction. Mol Gen Genet 212: 20–26 (1988).
Back E, Dunne W, Schneiderbauer A, de Framond A, Rastogi R, Rothstein SJ: Isolation of the spinach nitrite reductase gene promoter which confers nitrate inducibility on GUS gene expression in transgenic tobacco. Plant Mol Biol 17: 9–18 (1991).
Becker TW, Foyer C, Caboche M: Light-regulated expression of the nitrate-reductase and nitrite-reductase genes in tomato and in the phytocrome-deficient aurea mutant of tomato. Planta 188: 39–47 (1992).
Bevan M: Binary Agrobacterium vectors for plant transformation. Nucl Acids Res 12: 8711–8721 (1984).
Boutry M, Chua N-H: A nuclear gene encoding the β-subunit of the mitochondrial ATP synthase in Nicotiana plumbaginifolia. EMBO J 4: 2159–2165 (1985).
Bradford M: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 (1976).
Brown JWS: A catalogue of splice junction and putative branch poin sequences from plant introns. Nucl Acids Res 14: 9549–9559 (1986).
Cheng C-L, Acedo GN, Christinsin M, Conkling MA: Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA 89: 1861–1864 (1992).
Dellaporta SL, Wood J, Hicks JB: A plant DNA minipreparation: Version II. Plant Mol Biol 1: 19–21 (1983).
Duncanson E, Gilkes AF, Kirk DW, Sherman A, Wray JL: Nirl, a conditional-lethal mutation in barley causing a defect in nitrite reduction. Mol Gen Genet 236: 275–282 (1993).
Faure J-D, Vincentz M, Kronenberger J, Caboche M: Co-regulated expression of nitrate and nitrite reductases. Plant J 1: 107–113 (1991).
Friemann A, Brinkmann K, Hachtel W: Sequence of a cDNA encoding nitrite reductase from the tree Betula pendula and identification of conserved protein regions. Mol Gen Genet 231: 411–416 (1992).
Friemann A, Lange M, Hachtel W, Brinkmann K: Induction of nitrate assimilatory enzymes in the tree Betula pendula. Plant Physiol 99: 837–842 (1992).
Hasumi H, Nagata E, Nakamura S: Molecular heterogeneity of ferredoxin-NADP+ reductase from spinach leaves. Biochem Biophys Res Commun 110: 280–286 (1983).
Hirasawa M, de Best JH, Knaff DB: The effect of lysine- and arginine-modifying reagents on spinach ferredoxin: nitrite oxidoreductase. Biochim Biophys Acta 1140: 304–312 (1993).
Hoff T, Stummann BM, Henningsen KW: Cloning and expression of a gene encoding a root specific nitrate reductase in bean (Phaseolus vulgaris). Physiol Plant 82: 197–204 (1991).
Horsch RB, Fry JW, Hoffman NL, Eicholtz D, Rogers SG, Fraley RJ: A simple and general method for transferring genes into plants. Science 227: 1229–1231 (1985).
Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Jensen PE, Hoff T, Møller MG, Stummann BM, Henningsen KW: Identification and characterization of a nitrate reductase gene from bean (Phaseolus vulgaris) containing four introns. Submitted.
Kavanagh TA, Jefferson RA, Bevan MW: Targeting a foreign protein to chloroplasts using fusions to the transit peptide of a chlorophyll a/b protein. Mol Gen Genet 215: 38–45 (1988).
Keegstra K, Olsen LJ, Theg SM: Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Physiol Plant Mol Biol 40: 471–501 (1989).
Kronenberger J, Lepingle A, Caboche M, Vaucheret H: Cloning and expression of distinct nitrite reductases in tobacco leaves and roots. Mol Gen Genet 236: 203–208 (1993).
Lahners K, Kramer V, Back E, Privalle L, Rothstein S: Molecular cloning of complementary DNA encoding maize nitrite reductase. Plant Physiol 88: 741–746 (1988).
Mohr H, Neininger A, Seith B: Control of nitrate reductase and nitrite reductase gene expression by light, nitrate and a plastidic factor. Bot Acta 105: 81–89 (1992).
Murashige T, Skoog F: A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 (1962).
Neininger A, Bichler J, Schneiderbauer A, Mohr H: Response of a nitrite-reductase 3.1-kilobase upstream regulatory sequence fro spinach to nitrate and light in transgenic tobacco. Planta 189: 440–442 (1993).
Neininger A, Kronenberger J, Mohr H: Coaction of light and a plastidic factor in controlling nitrite-reductase gene expression in tobacco. Planta 187: 381–387 (1992).
Oelmuller R, Schuster C, Mohr H: Physiological characterization of a plastidic signal required for nitrate induced appearance of nitrate and nitrite reductases. Planta 174: 75–83 (1988).
Rastogi R, Back E, Schneiderbauer A, Bowsher CG, Moffat B, Rothstein SJ: A 330 bp region of the spinach nitrite reductase gene promoter directs nitrate-inducible tissue-specific expression in transgenic tobacco. Plant J 4: 317–326 (1993).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Schuster C, Mohr H: Appearance of nitrite-reductase mRNA in mustard seedling cotyledons is regulated by phytocrome. Planta 181: 327–334 (1990).
Seith B, Schuster C, Mohr H: Coaction of light, nitrate and a plastidic factor in controlling nitrite-reductase gene expression in spinach. Planta 184: 74–80 (1991).
Tsay Y-F, Schroeder JI, Feldmann KA, Crawford NM: The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 (1993).
Vaucheret H, Kronenberger J, Lepingle A, Vilaine F, Boutin J-P, Caboche M: Inhibition of tobacco nitrite reductase activity by expression of antisense RNA. Plant J 2: 559–569 (1992).
Vincentz M, Moureaux T, Leydecker M-T, Vaucheret H, Caboche M: Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J 3: 315–324 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sander, L., Jensen, P.E., Back, L.F. et al. Structure and expression of a nitrite reductase gene from bean (Phaseolus vulgaris) and promoter analysis in transgenic tobacco. Plant Mol Biol 27, 165–177 (1995). https://doi.org/10.1007/BF00019188
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00019188