Abstract
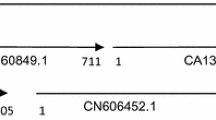
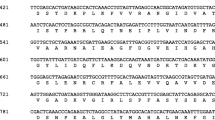
Clones representing two distinct barley catalase genes, Cat1 and Cat2, were found in a cDNA library prepared from seedling polysomal mRNA. Both clones were sequenced, and their deduced amino acid sequences were found to have high homology with maize and rice catalase genes. Cat1 had a 91% deduced amino acid sequence identity to CAT-1 of maize and 92% to CAT B of rice. Cat2 had 72 and 79% amino acid sequence identities to maize CAT-2 and-3 and 89% to CAT A of rice. Barley, maize or rice isozymes could be divided into two distinct groups by amino acid homologies, with one group homologous to the mitochondria-associated CAT-3 of maize and the other homologous to the maize peroxisomal/glyoxysomal CAT-1. Both barley CATs contained possible peroxisomal targeting signals, but neither had favorable mitochondrial targeting sequences. Cat1 mRNA occurred in whole endosperms (aleurones plus starchy endosperm), in isolated aleurones and in developing seeds, but Cat2 mRNA was virtually absent. Both mRNAs displayed different developmental expression patterns in scutella of germinating seeds. Cat2 mRNA predominated in etiolated seedling shoots and leaf blades. Barley genomic DNA contained two genes for Cat1 and one gene for Cat2. The Cat2 gene was mapped to the long arm of chromosome 4, 2.9 cM in telomeric orientation from the mlo locus conferring resistance to the powdery mildew fungus (Erysiphe graminis f.sp. hordei).
Similar content being viewed by others
References
Benito C, Figuereiras AM, Gonzalez MT, Salinas J: Biochemical evidence of homoeology between wheat and barley chromosomes. Z Pflanzenzüchtg 94: 307–320 (1985).
Bethards LA, Skadsen RS, Scandalios JG: Isolation and characterization of a cDNA clone for the Cat2 gene in maize and its homology with other catalases. Proc Natl Acad Sci USA 84: 6830–6834 (1987).
Bietz JA: Cereal prolamin evolution and homology revealed by sequence analysis. Biochem Genet 20: 1039–1053 (1982).
Chen Z, Silva H, Klessig DF: Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262: 1883–1886 (1993).
Chirgwin JM, Prybyla AE, Mac Donald RJ, Rutter WJ: Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18: 5294–5299 (1979).
Cohen G, Fessl F, Traczyk A, Rytka J, Ruis H: Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol Gen Genet 200: 74–79 (1985).
Fita I, Rossmann MG: The active center of catalase. J Mol Biol 185: 21–37 (1985).
GCG Program Manual for the Wisconsin Package, Version 8, Aug., 1994. Genetics Computer Group, 575 Science Dr., Madison, WI 53711, USA.
Gebhardt C, Ritter E, Debener T, Schachtschabel U, Walkemeier B, Uhrig H, Salamini F: RFLP analysis and linkage mapping in Solanum tuberosum. Theor Appl Genet 78: 65–75 (1989).
Gietl C, Faber KN, van derKleim IJ, Veenhuis M: Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci USA 91: 3151–3155 (1994).
Gonzalez E: The C-terminal domain of plant catalase: Implication for a glyoxysomal targeting sequence. Eur J Biochem 199: 211–215 (1991).
Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S: A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol 108: 1657–1664 (1989).
Hartl F-U, Pfanner N, Nicholson DW, Neupert W: Mitochondrial protein import. Biochim Biophys Acta 988: 1–45 (1989).
Havir EA, McHale NA: Enhanced-peroxidatic activity in specific catalase isozymes of tobacco, barley, and maize. Plant Physiol 91: 812–815 (1989).
Hinze K, Thompson RD, Ritter E, Salamini F, Schulze-Lefert P: RFLP-mediated targeting of the mlo resistance locus in barley (Hordeum vulgare). Proc Natl Acad Sci USA 88: 3691–3695 (1991).
Joshi CP: Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucl Acids Res 15: 9627–9640 (1987).
Kendall AC, Keys AJ, Turner JC, Lea PJ, Mifflin B: The isolation and characterization of a catalase-deficient mutant of barley (Hordeum vulgare L.). Planta 159: 505–511 (1983).
Kleinhofs A, Kilian A, Saghai MA, et al.: A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet 86: 705–712 (1993).
Koo JC, Chun HJ, Ha MS, Choi SO, Lee WS, Hong JC, Bahk JD, Allen RD, Cho MJ: Characterization of putative peroxisomal targeting signals of cotton catalase. Abstract 1236, 4th International Congress of Plant Molecular Biology, Amsterdam, June, 1994.
Kozak M: Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 (1986).
Levine A, Tenhaken R, Dixon R, Lamb C: H202 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 (1994).
Loewen PC, Triggs BL: Genetic mapping of katF, a locus what with katE affects the synthesis of a second catalase species in Escherichia coli. J Bact 160: 668–675 (1984).
Mori H, Higo K, Minobe Y, Matsui H, Chiba S: Nucleotide and derived amino acid sequence of a catalase cDNA isolated from rice immature seeds. Plant Mol Biol 18: 973–976 (1992).
Morita S, Tasaka M, Fujisawa H, Ushimaru T, Tsuji H: A cDNA clone encoding a rice catalase isozyme. Plant Physiol 105: 1015–1016 (1994).
Ossowski Iv, Hausner G, Loewen PC: Molecular evolutionary analysis based on the amino acid sequence of catalase. J Mol Evol 37: 71–76 (1993).
Patterson BD, Payne LA, Chen Y-Z, Graham D: An inhibitor of catalase induced by cold in chilling-sensitive plants. Plant Physiol 76: 1014–1018 (1984).
Prasad TK, Anderson MD, Stewart CR: Acclimation, hydrogen peroxide, and abscisic acid protect mitochondria against irreversible chilling injury in maize seedlings. Plant Physiol 105: 619–627 (1994).
Redinbaugh MG, Sabre JG, Scandalios JG: Expression of the maize Cat3 catalase gene is under the influence of a circadian rhythm. Proc Natl Acad Sci USA 87: 6853–6857 (1990).
Redinbaugh MG, Wadsworth GJ, Scandalios JG: Characterization of catalase transcripts and their differential expression in maize. Biochim Biophys Acta 951: 104–116 (1988).
Sanger F, Nicklen S, Coulson AR: DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 (1977).
Scandalios JG: Regulation and properties of plant catalases. In: Foyer CH, Mullineaux PM (eds) Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants, pp. 275–315. CRC Press, Boca Raton, FL (1994).
Scandalios JG, Tsaftaris AS, Chandlee JM, Skadsen RW: Expression of the developmentally regulated catalase (Cat) genes in maize. Devel Genet 4: 281–293 (1984).
Skadsen RW, Tibbot BK: Temporal expression patterns of alpha-amylase isozymal genes in polysomal and total RNAs of germinating barleys. J Cereal Sci 19: 199–208 (1994).
Skadsen RW: Aleurones from a barley with low α-amylase activity become highly responsive to gibberellin when detached from the starchy endosperm. Plant Physiol 102: 195–203 (1993).
Skadsen RW, Scandalios JG: Translational control of photo-induced expression of the Cat2 catalase gene during leaf development in maize. Proc Natl Acad Sci USA 84: 2785–2789 (1987).
Spevak W, Fessl F, Rytka J, Traczyk A, Skoneczny M, Ruis H: Isolation of the catalase T structural gene of Saccharomyces cerevisiae by functional complementation. Mol Cell Biol 3: 1545–1551 (1983).
Suzuki M, Ario T, Hattori T, Nakamura K, Asahi T: Isolation and characterization of two tightly linked catalase genes from castor bean that are differentially regulated. Plant Mol Biol 25: 507–516 (1994).
Tsaftaris AS, Bosabalisis AM, Scandalios JG: Cell-type-specific gene expression and acatalasemic peroxisomes in a null Cat2 catalase mutant of maize. Proc Natl Acad Sci USA 80: 4455–4459 (1983).
Wettstein-Knowles Pv: Clones and mapped genes: current status. In: Barley Shewry PR, (ed) Genetics, Biochemistry, Molecular Biology and Biotechnology, Biotechnology in Agriculture No. 5, pp. 73–98. CAB International, Wallingford, UK (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Skadsen, R.W., Schulze-Lefert, P. & Herbst, J.M. Molecular cloning, characterization and expression analysis of two catalase isozyme genes in barley. Plant Mol Biol 29, 1005–1014 (1995). https://doi.org/10.1007/BF00014973
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00014973