Abstract
Purpose
The first step in typical treatment of vestibular schwannoma (VS) is to localize the tumor region, which is time-consuming and subjective because it relies on repeatedly reviewing different parametric magnetic resonance (MR) images. A reliable, automatic VS detection method can streamline the process.
Methods
A convolutional neural network architecture, namely YOLO-v2 with a residual network as a backbone, was used to detect VS tumors from MR images. To heighten performance, T1-weighted–contrast-enhanced, T2-weighted, and T1-weighted images were combined into triple-channel images for feature learning. The triple-channel images were cropped into three sizes to serve as input images of YOLO-v2. The VS detection effectiveness levels were evaluated for two backbone residual networks that downsampled the inputs by 16 and 32.
Results
The results demonstrated the VS detection capability of YOLO-v2 with a residual network as a backbone model. The average precision was 0.7953 for a model with 416 × 416-pixel input images and 16 instances of downsampling, when both the thresholds of confidence score and intersection-over-union were set to 0.5. In addition, under an appropriate threshold of confidence score, a high average precision, namely 0.8171, was attained by using a model with 448 × 448-pixel input images and 16 instances of downsampling.
Conclusion
We demonstrated successful VS tumor detection by using a YOLO-v2 with a residual network as a backbone model on resized triple-parametric MR images. The results indicated the influence of image size, downsampling strategy, and confidence score threshold on VS tumor detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Vestibular schwannoma (VS), also named acoustic neuroma, is a benign intracranial tumor. If such a tumor grows, it might compress the brainstem and cerebellum and might cause hearing impairment, tinnitus, dizziness, syncope, trigeminal neuropathy, and facial palsy. Various treatments can be applied for VS; of these, Gamma Knife radiosurgery (GKRS) is a radiosurgery technique usually applied to small- and medium-sized (<2.5-cm) VS tumors. In addition, magnetic resonance (MR) images scanned in terms of different parameters that provide high-contrast images for particular tissues play a pivotal role in diagnosis and treatment planning. By visually inspecting these parametric MR images, physicians first detect the tumor lesions, delineate the tumor contour and subsequently determine the VS treatment [1]. MR images of different parameters can be used to inspect the two main types of VS lesions: uniform solid tumor parts with enhancement on T1-weighted (T1W)–gadolinium contrast-enhanced (T1W+C) images and cystic parts with enhancement on T2-weighted (T2W) images. During GKRS planning, tumor localization is the first step; it relies on experienced neurosurgeons and neuroradiologists repeatedly reviewing different parametric MR images. Hence, it is time-consuming and subjective. A reliable automatic VS detection can increase the efficiency of GKRS planning.
Scholars have published numerous deep learning methods that have been successfully applied to medical images; convolutional neural networks (CNN) have been especially effective. Such networks produce effective feature maps performing comparably with predefined features used for image recognition [2, 3]. Some CNN-based semantic segmentation algorithms, such as fully convolutional networks (FCNs) [4] and U-Nets [5], form a standard approach for detection in the clinical context, and substantially assist with clinical challenges like radiotherapeutic planning [6,7,8,9].
These studies displayed that the pixel-wise predictions helped to localize brain tumors precisely, which might enhance tumor volume measurements and treatment response determination. However, object detection networks usually estimate whether objects exist in a region; thus, they may provide features that are not relevant to the semantic segmentation but are highly relevant to the relationship between the region and the background [9]. Object detection techniques have been widely used in previous work in other medical images. For breast cancer detection, Li et al. [10] demonstrated automatic detection thyroid papillary cancer in ultrasound images by using faster RCNN. Mohammed et al. [11] detected breast masses in digital mammograms using a YOLO-based computer aid diagnosis system. For chest testing, George et al. [12] used YOLO for real-time detection of lung nodules from low-dose CT scans. For skin testing, Ünver et al. [13] combined YOLO and GrabCut algorithm for segmentation of the skin lesion areas in dermoscopic images. These studies have demonstrated the strength of CNN object detection algorithms in the medical image domain.
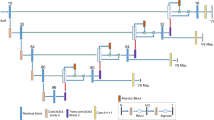
In addition, studies have demonstrated that using multiparametric MR images improves detection for brain tumors with multiple subregions [14,15,16]. For VS tumor detection, Wang et al. [17] and Shapey et al. [18] demonstrated the feasibility of CNN and multiparametric MR images to the segmentation of VS lesions. Lee et al. [19] improved VS lesion detection by treating multiparametric MR images from the same patient as different channels of the same image. These three related studies of VS detection used UNet-based semantic segmentation algorithms for pixel-wisely detecting the VS tumor lesions. Instead, the VS tumor as a whole can be detected within a region by using the proposed method, which provides an alternative tool for assisting clinician in localizing the tumor. The present study was to implement a region-wise method for VS tumor detection. We applied a CNN detection subnetwork, namely YOLO-v2, to automatically detect VS lesions using triple-channel images composed of T1W+C, T2W, and T1W images, and discussed the applicability under clinical circumstances. As the recognized object detection method, the YOLO-v2 provided a faster training duration and acceptable detection performance, so we can test the influence from different down-sampling strategies efficiently. We added a YOLO-v2 detection subnetwork after a residual network which is a stack of several different forms of residual convolution blocks. Such a deep architecture allows us to assess the influence of different features, in which the size varies from 13 x 13 pixels to 32 x 32 pixels, obtained from the residual convolution blocks. The YOLO-v2 block and residual networks were displayed in Fig. 2b. These feature-extraction layers produced feature maps that were downsampled by factors of 16 and 32. Also, before the training phase, we cropped the images for better focus on the brain. The detection capabilities of each CNN architecture were evaluated with average precision and F1 scores.
2 Materials and Methods
2.1 Data Acquisition and Preprocessing
During GKRS planning, the physicians locate the tumor regions to determine the target area and dose delivery. So that, the data of each VS patient typically contain not only the multiparametric MR images but also the individual annotations of the tumor region, which are manually marked by experienced neurosurgeons and neuroradiologists. Thus, we used these annotations as ground truth to formalize and to evaluate detection models.
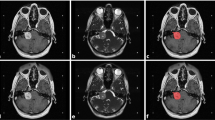
To extract adequate tumor features, the MR images with different parameters were required. In VS treatment, physicians focus on not only the solid part of a tumor but also the cystic part, and the two parts of the tumor are enhanced by T1W+C and T2W, respectively. Thus, we mainly used T1W+C and T2W images to detect the target region. Also, T1W images were used because adding T1W to the training phase had made a slight improvement over using only T1W+C and T2W in the previous study [19]. In this study, we retrospectively collected 516 pre-GKRS VS patients’ three-dimensional MR images and tumor regions from Taipei Veterans General Hospital, Taiwan. The study was approved by the Institutional Review Board of Taipei Veterans General Hospital (IRB-TPEVGH No.: 2018-11-0089AC). The MR images were completely anonymized. All the MR images were scanned by a GE scanner, magnetic field strength 1.5 Tesla, including T1W, T2W, and T1W+C axial two-dimensional Spin Echo images (Fig. 1a). The pixel matrix size of the MR images was 512 × 512 × 20 with a voxel size of 0.5 × 0.5 × 3 mm3 (in x–y–z directions; Fig. 1b). For the preprocessing, ANTs N4 bias field correction was applied to all images [20]. Because the individual tumor bounding boxes were manually annotated for T1W+C images by experienced neuroradiologists, T1W and T2W images were then respectively co-registrated to the corresponding individual T1W+C image using the rigid-body 6-degrees of freedom (6DOF) registration procedure by SPM12 (Statistic Parametric Mapping) [21]. These three parametric images were concurrently used as triple-channel input images for training models (Fig. 1c). A multichannel image (e.g. red–green–blue image) contains more information than a single-channel image (e.g. grayscale) because features can be either within-channel features or cross-channel features. In this study, we treated the 512 × 512 × 20 data as 20 slices of 512 × 512 images because we used two-dimensional object detectors to detect the VS tumors, and the data provided a higher amount of information in the x–y plane (axial view). We randomly separated the preprocessed image data into 412 subjects as the training set (80% of all subjects) and 104 subjects as the evaluation set (20% of all subjects). Because we applied two-dimensional object detection methods to devise tumor detection models, the data of each patient included 20 triple-channel images in the axial view with matrix 512 × 512. During the training phase, only the images that contain tumors would be used in the following step of model training. The amount of 2342 two-dimensional slices marked with tumors were selected as training inputs. During the evaluation phase, a total of 2080 images with or without tumor markers were utilized for evaluating the model. Among the set for evaluation, 609 images were marked with tumors, including 618 different tumor volumes.
(a) Imaging of VS on different parametric MR images. The solid part of tumors appears with high intensity in T1W+C (left). The cystic part appears with high intensity in T2W (right). The T1W (middle) are used for additional image information. (b) The MR image data are in matrix size 512 × 512 × 20. (c) Three parametric MR images are used as the triple-channel images
2.2 Experimental Procedure
In the research of deep-learning architecture for object detection, there are two commonly used strategies: one-stage and two-stage strategies. The first type was a two-stage detector, using a Region Proposal Network that localized objects from backgrounds and then sent the region proposals to the subsequent networks for classification and regression. The second type was a one-stage detector, using methods such as YOLO algorithms, treating object detection as a regression problem by designating a single network to determine the class probabilities and bounding box locations simultaneously. A previous study demonstrated that a one-stage detector produced the prediction directly from the corresponding anchors in the feature map, generating a faster detection [22,23,24,25].
In the present study, we applied two CNN-based architectures, YOLO-v2 with deep residual network as a backbone model for VS detection, where the backbone model was pretrained as a feature extractor [23, 24]. For such deep models, the input images must be downsampled to the feature maps through the extraction of multilayers. A previous study demonstrated superior detection performance levels by downsampling the input images by a factor of 32 [24]. Accordingly, we applied architectures that downsampled the input images by a factor of 32, and applied another factor of 16, as depicted in Fig. 2a. In addition, the aforementioned architectures used sets of predefined anchor boxes to improve the effectiveness of detection. The anchor boxes were the initial predictive guesses regarding the bounding boxes; the sizes were determined based on the specific object sizes of the training data sets. During detection, the anchor boxes tiled across the images or feature maps so the networks could predict the probabilities and refine the corresponding anchor boxes, instead of predicting bounding boxes directly. However, too many anchor boxes may increase the computation cost and lead to overfitting. Therefore, the number of anchor boxes is an influential hyperparameter for detectors. Rather than hand-picking, we used an approximate k-means algorithm to determine the sizes of anchor boxes by clustering the intersection-over-union (IoU) distance between each pair of ground truth bounding boxes [24]. The IoU and IoU distance (DIoU) are defined as:
Flow of the present study. (a) Data preparation and preprocessing: we converted the images to 416 × 416 or 448 × 448 by cropping from original 512 × 512, and constructed triple-channel images by T1W+C, T1W, and T2W MR images. Data with these three image sizes were trained and tested separately. (b) Training phase of detection models: training the YOLO-v2 detectors using the preprocessed images. The residual network was a stack of convolution blocks as displayed. The N was set to 2 and 3, which would downsampled the input by 16 and 32, respectively. (c) Evaluation phase for detection models by average precision and F1 score
where A represents the ground truth bounding box, and B represents the predicted bounding box. We ran k-means algorithms with various values of k, where k represents the number of anchor boxes; we examined the tradeoff between the number of anchor boxes and the mean of IoU, and further determined the appropriate k.
2.3 Cropping Images
Each network architecture has its own specific suitable or restricted input size, so the result of detection is influenced by the size of the input images. Hence, before entering the training phase of the models, we cropped the triple-channel images as training data in addition to the 512 × 512 raw images. We inferred that cropping images allows the model to focus better on the brain without the backgrounds in the image, with a view to improving detection accuracy and speeding up the training process. Because the model downsampled the input images by a factor of 32, we cropped the images around the brain part to 416 × 416 pixels, which are the multiples of 32. We created the brain masks of the images by the following steps: First, we computed a global image threshold using Otsu’s method, which determines a threshold by minimizing intraclass intensity variance [26]. The original grayscale images were then converted to binary images. Subsequently, filling the holes in the binary images, where the holes were sets of black pixels that cannot be reached from the edges of the binary images. Then, we conducted an area opening operation on the binary images. Thus, we gained whole-brain masks from the original 512 × 512-pixel MR images. Subsequently, we cropped the original images to 416 × 416 or 448 x 448, centered on the central points of brain masks.
2.4 Evaluation of Each Tumor Detection Model
The main goal of the present study was to explore tumor detection effects through different pipelines and models, instead of demonstrating the regression error between bounding boxes. Three types of performance evaluations for single-class object detection according to the ground truth and prediction were used in the present study: true positive (TP), false positive (FP), and false negative (FN). The FP indicated predicted bounding boxes without any tumor, and the TP or FN were determined by whether the IoU between the prediction and the ground truth bounding boxes was greater than a predefined IoU threshold set to 0.5. With the TP, FP, and FN, we calculated recall, precision, and F1 scores. Precision represents how accurate the predictions are; recall represents how accurately the model finds all the positive items. However, evaluating a model using only recall or precision may produce bias. Therefore, the F1 score calculated by the harmonic mean of the precision and recall was estimated for better comprehension [27].
In addition to the location, the confidence score of each predicted bounding box was another crucial value for detection performance measurement. During the evaluation phase, the confidence score of each bounding box represents the model's confidence in the detection, and it was one of the outputs of the neural network using regression. Through adjustments for the lower limit of the threshold of confidence score, a different number of bounding boxes would remain. Reducing the threshold would retain more bounding boxes, which resulted in the higher recall but lower precision. The appropriate threshold of confidence score for the model was selected by exploring out the highest F1 score under the specific threshold of confidence score. In addition, the precision–recall curve (P-R curve) was drawn by utilizing the precision and recall values calculated under the different thresholds of confidence score. Then, the average precision (AP) was estimated by calculating the area under the P-R curve [28]. The performance levels of the models were evaluated by the APs and F1 scores [29, 30].
The codes were implemented by using MATLAB [31] and executed with an NVIDIA GeForce RTX 2080 Ti GPU graphics card (NVIDIA, Santa Clara, CA), an Intel Core i7-8700 CPU, and a 32 GB RAM.
3 Results
In this section, we first demonstrate a relationship between mean IoU and various numbers of anchor boxes, which provided with a reference to select a suitable number of anchor boxes. Then, we list the recall, precision, F1 score, and AP of each model with different input sizes and different downscaling factors to evaluate the detection capabilities under different confidence score thresholds.
3.1 Number of Anchor Boxes
Because using too many anchor boxes would result in time-consuming and inaccurate detection attempts, we did not choose the number of anchor boxes, referred to as k, by the highest mean IoU, but by the tradeoff between number of anchor boxes and mean IoU. We implemented k-means with various values of k and plotted the tradeoff between number of anchor boxes and mean IoU (Fig. 3). As illustrated in Fig. 3a, we assumed that using six and seven anchor boxes, whose values of mean IoU were 0.7332 and 0.7965, respectively, would result in better detection, because using 10 or more anchor boxes only yield slightly higher mean IoU. However, the mean IoU and the size variation of anchor boxes were similar at these two numbers of anchors. To generate highly effective performance, we selected six predefined anchor boxes for training our VS detectors.
k-Means clustering was used for choosing the bounding boxes in our training data set to obtain prior anchor boxes. (a) The changes in the mean IoU with various number of anchor boxes k. The problem has a tradeoff between mean IoU and number of anchor boxes when k = 6 or 7. (b) The size of relative anchor boxes when k = 6 and 7. The size variation of anchor boxes is similar under the two conditions
3.2 AP of VS Detection with Different Input Sizes and Different Downscaling Factors
The AP presented in Table 1 demonstrates the detection capability of each combination of downsampling factors and input sizes. Separation of the brain region from background in the MR images may enhance the capability of detecting tumors because fewer excessive areas require examination; accordingly, the AP of the original input size, 512 × 512, was the worst among the models with the same downsampling strategy (Tables 1 and 2). In addition, a study used a model that downsampled the input images by a factor of 32 and gained desirable results on object detection benchmark data sets [24]. However, in our VS detection with triple-parametric MR images, the models that downsampled the input images by a factor of 16 achieved better performance levels than that by a factor of 32, regardless of how the input images were resized. The P-R curves and APs of each model with different input sizes and downscaling factors are shown in Fig. 4.
P-R curve of each model. The blue curves represent the performance of VS detection with the images cropped to 416 × 416, and the red curves and the yellow curves represent that the images cropped to the 448 × 448 size and the original 512 × 512 size, respectively. The black * denotes the precision and recall when the threshold of confidence score was set to 0.5
3.3 F1 Scores of VS Detection Under Various Threshold of Confidence Score
Next, we demonstrated the detection capabilities under the different thresholds of confidence score. When the confidence score was set to 0.5, the model combined with 416 × 416-pixel input and downsampling by 16 attained the best F1 score at 0.7953 among the models with the same downsampling strategy. However, the model combined with 448 × 448-pixel input and downsampling by 16 attained a lower F1 score of 0.7730 but a higher recall of 0.7880, which means that more tumors were detected although the tumor prediction proposal was less trustworthy. Moreover, the results presented in Table 1 and Fig. 5 proved that if an appropriate threshold of confidence score was selected, such as 0.56, the model combined with 448 × 448-pixel input and downsampling by 16 generated the highest F1 score, namely 0.8171. It indicated that the threshold of confidence score influenced the performance of detectors measurably.
F1 score of each detector trained with different input sizes and different downscaling factors. The models that downsampled the input images by 16 were outperformed the models that downsampled the input images by 32. The 416 × 416-pixel images with 16 instances of downsampling performed the best (F1 score 0.7953) at the threshold of confidence score set to 0.5. However, by adjusting the threshold of confidence to slightly higher value, the performance of each model attained better performance, and the 416 × 416-pixel images with downsampling by 16 performed the best (the highest F1 score 0.8171)
4 Discussion
To our knowledge, this is the first study that demonstrated VS tumor detection capability by models contained different combinations of input-image sizes and downsampling strategies from triple-parametric MR images. In the context of medical image detection, these detected areas were selected primarily to provide physicians with a preliminary examination that would allow them to screen out the location of VS quickly and to establish a follow-up treatment process. The present study proved that the YOLO-v2 with residual network as backbone model successfully detected VS tumors. Among the models, the 416 × 416-pixel input-image architecture that downsampled the input by 16 was the architecture that attained the best AP, namely 0.779, when the threshold of confidence was set to 0.5. The results suggested that removing the unrelated background increased the detection capability, and demonstrated that too much downsampling was unsuitable for detection. It can be inferred that deeper models do not always produce better performance.
Moreover, the P-R curve and F1 scores under different confidence score thresholds demonstrated the influence of the threshold selection for confidence score on the model performance. The model with the threshold of confidence score set at 0.5 in the present study did not obtain the best performance. However, the threshold of confidence score is not a learnable parameter that could be computed through the training process. We can only select a relatively suitable value by additional validations, which would be necessary if a practical detection model for clinical use were required.
Furthermore, we compared the YOLO-v2 and Faster RCNN algorithms, both with the 16-times downsampling feature extraction network, and found that the Faster RCNN requires a much larger memory. More specifically, the region proposal network in the Faster RCNN outputs regional images of various sizes and this process acquires a lot of memory. Besides, in the training phase, YOLO-v2 can process 16 images per batch whereas the Faster RCNN can only process 1 image per batch. In addition, the Faster RCNN took more than eight hours to train 5 epochs, while the YOLOv2 took less than 20 minutes to train 10 epochs. This is due to the limited GPU RAM of our equipment, which cannot accommodate large image for processing the Faster RCNN. Therefore, we adopted the YOLO-v2 since it provided a faster training duration and superior performance (AP = 0.6654 with 416 × 416-pixel input images, and AP = 0.7077 with 448 × 448-pixel input images) in this study.
In addition, the processing was practical in the present study because most of clinical MR images of VS have lower through-plane resolution relative to in-plane (anisotropic voxel size) resolution. We used two-dimensional axial view (x-y plane) information instead of three-dimensional data from each subject. The dimension of the through-plane (z direction) of the data was only 20 in the present study; the feature map would vanish in the process of extraction by multiple convolution layers. Therefore, with the current detection method, two-dimensional information can convey more meaningful characteristics for detection. However, three-dimensional data provide more spatial information, which may aid in individual VS detection. Such speculation warrants future investigations to produce additional evidence.
5 Conclusion
The contributions of the present study included two aspects: First, we demonstrated a YOLO-v2 with a residual network as a backbone that successfully detected VS tumors by resized triple-parametric MR images. Second, we implemented various resized images and architectures that downsampled input by different factors and gained an AP of 0.7953 using 416 × 416-pixel input images and downsampling by 16 when the thresholds of both confidence score and IoU were set to 0.5. In addition, although the two-dimensional detectors provided accurate performance on VS detection, a three-dimensional detector should be developed to evaluate how effectiveness can be enhanced for individual VS detection.
References
Wu, C.-C., Guo, W.-Y., Chung, W.-Y., Wu, H.-M., Lin, C.-J., Lee, C.-C., Liu, K.-D., & Yang, H.-C. (2017). Magnetic resonance imaging characteristics and the prediction of outcome of vestibular schwannomas following Gamma Knife radiosurgery. Journal of neurosurgery, 127, 1384–1391.
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems, 25, 1097–105.
LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998). Gradient-based learning applied to document recognition. Proc IEEE, 86, 2278–324.
Long J, Shelhamer E, Darrell T. 2015 Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 3431–40.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: convolutional networks for biomedical image segmentation. In Nassir Navab, Joachim Hornegger, William M. Wells, & Alejandro F. Frangi (Eds.), International conference on medical image computing and computer-assisted intervention (pp. 234–41). Cham: Springer.
de Brebisson A, Montana G. 2015 Deep neural networks for anatomical brain segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 20–8.
Dolz, J., Desrosiers, C., & Ayed, I. B. (2018). 3D fully convolutional networks for subcortical segmentation in MRI: a large-scale study. NeuroImage, 170, 456–70.
Pereira, S., Pinto, A., Alves, V., & Silva, C. A. (2016). Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Transactions on Medical Imaging, 35, 1240–51.
Jaeger, P. F., Kohl, S. A., Bickelhaupt, S., Isensee, F., Kuder, T. A., Schlemmer, H. P., Maier-Hein, K. H. (2020). Retina U-Net: Embarrassingly simple exploitation of segmentation supervision for medical object detection. In Machine Learning for Health Workshop (pp. 171-183).
Li, H., Weng, J., Shi, Y., Gu, W., Mao, Y., Wang, Y., & Zhang, J. (2018). An improved deep learning approach for detection of thyroid papillary cancer in ultrasound images. Scientific reports, 8(1), 1–12.
Al-Masni, M. A., Al-Antari, M. A., Park, J. M., Gi, G., Kim, T. Y., Rivera, P., Valarezo, E., Choi, M.-T., HanS-M, Kim, & T. S. . (2018). Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Computer methods and programs in biomedicine, 157, 85–94.
George, J., Skaria, S., & Varun, V. V. (2018). Using YOLO based deep learning network for real time detection and localization of lung nodules from low dose CT scans. In P. Nicholas & M. Kensaku (Eds.), Medical Imaging 2018 Computer-Aided Diagnosis, 10575 (p. 105751I). Washington: International Society for Optics and Photonics.
Ünver, H. M., & Ayan, E. (2019). Skin lesion segmentation in dermoscopic images with combination of YOLO and grabcut algorithm. Diagnostics, 9(3), 72.
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J., Freymann, J., Farahani, K., & Davatzikos, C. (2017). Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., & Wiest, R. (2014). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging, 34, 1993–2024.
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J. S., Freymann, J. B., Farahani, K., & Davatzikos, C. (2017). Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific data, 4, 170117.
Wang, G., Shapey, J., Li, W., Dorent, R., Demitriadis, A., Bisdas, S., Vercauteren, T., et al. (2019). Automatic segmentation of vestibular schwannoma from T2-weighted MRI by deep spatial attention with hardness-weighted loss. International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 264–272). Cham: Springer.
Shapey, J., Wang, G., Dorent, R., Dimitriadis, A., Li, W., Paddick, I., Bradford, R., et al. (2019). An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRI. Journal of Neurosurgery. https://doi.org/10.3171/2019.9.JNS191949
Lee, W. K., Wu, C. C., Lee, C. C., Lu, C. F., Yang, H. C., Wu, Y. T., Guo, W. Y., et al. (2020). Combining analysis of multi-parametric MR images into a convolutional neural network: Precise target delineation for vestibular schwannoma treatment planning. Artificial Intelligence in Medicine, 107, 101911.
Ashburner, John. (2012). SPM: A history. Neuroimage, 62(2), 791–800.
Tustison, N. J., Avants, B. B., Cook, P. A., Zheng, Y., Egan, A., Yushkevich, P. A., & Gee, J. C. (2010). N4ITK: improved N3 bias correction. IEEE transactions on medical imaging, 29, 1310.
Ren, Shaoqing, He, Kaiming, Girshick, Ross, & Sun, Jian. (2015). Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. Advances in Neural Information Processing Systems. https://doi.org/10.1109/TPAMI.2016.2577031
Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. 2016 Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778
Redmon, Joseph, and Ali Farhadi. "YOLO9000: Better, Faster, Stronger." 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, 2017.
Soviany, P., & Ionescu, R. T. (2018, September). Optimizing the trade-off between single-stage and two-stage deep object detectors using image difficulty prediction. In 2018 20th International Symposium on Symbolic and Numeric Algorithms for Scientific Computing (SYNASC). IEEE, pp. 209-214
Otsu, N. (1979). A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics, 9(1), 62–66.
Zhu, M. (2004). Recall, precision and average precision. Department of Statistics and Actuarial Science, University of Waterloo, Waterloo, 2, 30.
Powers, D. M. (2011). Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation.
Csurka, G., D. Larlus, and F. Perronnin. 2013 "What is a good evaluation measure for semantic segmentation?" Proceedings of the British Machine Vision Conference, pp. 32.1–32.11.
Everingham, M., Eslami, S. A., Van Gool, L., Williams, C. K., Winn, J., & Zisserman, A. (2015). The pascal visual object classes challenge: A retrospective. International Journal of Computer Vision, 111(1), 98–136.
The MathWorks, Inc. 2020 MATLAB Deep Learning Toolbox_User's Guide-The MathWorks, Inc.
Acknowledgements
The authors gratefully acknowledge the financial support in part by the Ministry of Science and Technology, Taiwan, R.O.C. (Grant No. MOST 108-2634-F-010-002 and No. MOST 110-2221-E-A49A-504-MY3), Veteran General Hospital University System of Taiwan (Grant No. VGHUST110-G7-2-1), and National Yang-Ming University (Grant No. 110BRC-B701). This manuscript was edited by Wallace Academic Editing.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, TH., Lee, WK., Wu, CC. et al. Detection of Vestibular Schwannoma on Triple-parametric Magnetic Resonance Images Using Convolutional Neural Networks. J. Med. Biol. Eng. 41, 626–635 (2021). https://doi.org/10.1007/s40846-021-00638-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-021-00638-8