Abstract
G-Quadruplexes (G4s) are non-canonical secondary structures formed within guanine-rich regions of DNA or RNA. G4 sequences/structures have been detected in human and in viral genomes, including Coronaviruses Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and SARS-CoV-2. Here, we outline the existing evidence indicating that G4 ligands and inhibitors of SARS-CoV-2 helicase may exert some antiviral activity reducing viral replication and can represent a potential therapeutic approach to tackle the COVID-19 pandemic due to SARS-CoV-2 infection. We also discuss how repositioning of FDA-approved drugs against helicase activity of other viruses, could represent a rapid strategy to limit deaths associated with COVID-19 pandemic.
Similar content being viewed by others
1 Severe Acute Respiratory Syndrome Coronavirus and the 2020 Pandemic
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) emerged for the first time in 2002 and 2003 in Guangdong province (China) and caused 800 deaths worldwide in 2003 [1]. In 2005, research conducted in different laboratories demonstrated that bats may be the natural reservoirs for SARS-CoV [2]. SARS-CoV is a positive single-stranded (ss) RNA virus with a large RNA genome of approximately 29.7 kb. SARS-CoV replicons consist of the genomic 5′ and 3′ untranslated regions (UTR) and an open reading frame (ORF) encoding for the polyprotein(pP)-a, and for pP-ab containing the sequence for 16 non-structural proteins (from Nsp1 to Nsp16) that constitute the replication/transcription complex of the virus [9]. The remaining part of the SARS-CoV genome encodes four essential structural proteins, including spike (S) glycoprotein, small envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein, and accessory proteins that may interfere with the host innate immune response [3].
In June 2012, the first case of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) was identified in a patient from the Kingdom of Saudi Arabia. MERS-CoV is a positive ssRNA virus with a genome of about 30.1 kb containing 10 predicted ORFs, 9 of which may be displayed from a set of nested 7 sub-genomic mRNA. ORF1a and ORF1b are the replicase opening frames which cover the 5′ end region of the genome, while the 3′ end encodes structural proteins, including S, E, M and N proteins, and several other accessory proteins [4].
In late 2019, several Chinese citizens from Wuhan experienced a pneumonia associated to SARS-CoV-2 infection, later designated by WHO as Coronavirus Disease 2019 (COVID-19) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). On 11 March 2020, WHO declared COVID-19 a “pandemic” condition. The figures of the COVID-19 pandemic are constantly and rapidly increasing throughout the globe. On 6 April 2020, according to WHO, the global number of confirmed infections is 1,174,866, while the death toll has topped 64,541 deaths (https://who.sprinklr.com/).
The clinical presentation of COVID-19 varies from asymptomatic or mild respiratory symptoms, to severe pneumonia with respiratory failure and death, mainly in elderly with other chronic underlying diseases [5]. To date, there are no available vaccines and there is very little evidence of the effectiveness of treatments that may reduce severity of disease and prevent death. As the global scientific community is in the race for finding novel therapies for the disease, in late April 2020 over 500 interventional clinical studies on the current COVID-19 have been registered at ClinicalTrials.gov [6]. While waiting for a preventative vaccine, most of the therapeutic strategies against COVID-19 are currently based on slowing down the virus replication (i.e. by using antivirals adopted for other viruses), and on reducing severe forms of disease (i.e. by counteracting the cytokine storm syndrome).
Most of the current studies are based on the similarity of the SARS-CoV-2 with SARS-CoV and MERS-CoV, even if the latter is the most genetically different from the novel coronavirus [7].
In fact, as reported by Zhou et al. [8], the SARS-CoV-2 shares a high nucleotide sequence identity with SARS-CoV (79.7%), revealing a conserved evolutionary relationship between these two viruses. Conceptually, the protein sequence and structure similarity of SARS-CoV-2 with other types of coronavirus might help to develop effective anti-COVID-19 approaches.
2 Biological Role of G-quadruplexes and their Impact as Potential Targets for COVID-19
G-quadruplexes (G4s) are tetrahelical structures formed by guanine-rich regions of DNA or RNA [9]. The building blocks of G4s are assembled in G-quartets that are connected by Hoogsteen hydrogen bonds to form polymorphic structures. In general, all G4 structures exhibit a favourable thermodynamics, even if RNA G4s are more stable than DNA G4s [10, 11]. Although the frequency of G4-forming sequences (G4FS) vary significantly among evolutionary groups, they have been identified in all species. In human genome, putative G4FS (PG4FS) were found in various regions including sub-telomeres, telomeres, gene bodies, and 5′ untranslated regions and other gene regulatory regions. This observation suggests that G4s are likely involved in the regulation of crucial biological pathways, such as replication and regulation of gene expression [9]. PG4FS sequences have been found not only in humans, but also in other mammalian genomes, bacteria, and, intriguingly, they were found also in viral genomes [12].
Ruggiero and Richter [13] reviewed the emerging evidence suggesting that PG4FS can be involved in viral replication and recombination, in virulence control, and in the regulation of other pivotal steps of viral life cycles. Moreover, a recent analysis of PG4FS in seven viral taxa (approximately available 7370 viral genome assemblies) infecting five distant eukaryotic hosts, suggested a co-evolution of virus and host G4s [14].
Lavezzo et al. [15] recently provided a list of PG4FS, reporting their position and statistical significance, and the degree of conservation among strains, in a pool of viruses following the Baltimore classification. Among the other viruses, the authors also reported the characteristics of PG4FS detected in the genome of several coronaviruses.
Interestingly, a multi-domain polypeptide in SARS-CoV, including the structurally organised domains, within the Nsp3, represents a SARS unique macrodomain that could bind G4s and play a key role in viral replication and transcription machinery [16, 17]. These findings suggest that targeting G4s in the viral genomes may lead to the exciting possibility of affecting replication of coronaviruses, including the recently emerged SARS-CoV-2.
Based on this evidence, we speculate that the use of G4-mediated antiviral drugs may represent a significant turning point in the therapeutic management of COVID-19. Several algorithms for detecting expected matching patterns for G4s formation have been already described [18].
Here, we used the Quadruplex forming G-Rich Sequences (QGRS) Mapper, a web-based server database [19], that generates detailed information on composition and distribution of PG4FS. Following QGRS method, the PG4FS in the genome of SARS-CoV-2 by Wuhan (NC_045512.2) were identified by the motif GxNy1GxNy2GxNy3Gx, where x was the number of G tetrads and y the length of loops connecting them. The criteria of the analysis included the setting of maximum length at 30 bases; a minimum G-group equal to 2; and loop size from 0 to 36.
In accordance with the findings by Lavezzo et al. in SARS-CoV [15], we identified 25 PG4FS in the SARS-CoV-2 genome (Table 1). This finding suggests that G4s ligands could represent a potential therapeutic strategy for COVID-19.
Although these data support the observation that G4s ligands may exert a reasonably antiviral activity, the use of these compounds in clinical trials is far from being adopted because of their low selectivity profile and poor drug-like properties.
3 Alternative Approaches to Target G4s in COVID-19
Interestingly, the therapeutic potential of G4s in coronavirus could be also reversed by molecules that block the activity of RNA helicases of the virus. This class of proteins can unwind G4s with a mechanism often required before virus or cellular machineries can act on the nucleic acid substrate, thus suggesting their important role in the virus replication [20].
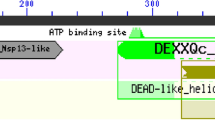
The nsp13 is a helicase that could unwind the 3′ end of the nascent RNA primer and facilitate transfer to the complementary region of the 5′-leader genomic sequence in SARS-CoV, thus regulating virus replication [21]. The functional relevance of helicases activities highlights that inhibitors or modulators for these enzymes are potentially important as therapeutic agents [22].
Tanner et al. [23] found that Bananin and three of their derivatives, exhibited activity as non-competitive inhibitors on nsp13 helicase, probably by an allosteric mechanism. Cho et al. [24], identified a novel small compound (7-ethyl-8-mercapto-3-methyl-3,7-dihydro-1H-purine-2,6-dione also designed as EMMDPD) able to inhibit ATP hydrolysis, as well as dsDNA unwinding activities of the SARS-CoV helicase, without any cell cytotoxicity.
Unfortunately, for many of these compounds, no further evidence exists on action mechanisms or potential translational application in clinical practice.
Interestingly, DNA aptamers, composed by two distinct classes of G4s and non-G4s forming sequences against the SARS-CoV helicase, were isolated using a SELEX (systematic evolution of ligands by exponential enrichment) approach [25]. These aptamers are oligonucleotides emerging as a class of molecules that may interact with viral components and interfere with viral replication [26].
While the identification of therapeutic aptamers requires further studies, we have to explore alternative plausible therapeutic strategies to face the emergency caused by COVID-19.
One possible alternative strategy is to inhibit viral helicase activity by using Food and Drug Administration (FDA)-approved drugs with scientific evidence of this molecular effect, with no toxic effects. Interestingly, after a search in the DrugBank database (https://www.drugbank.ca/, version 5.1.5, released 2020-01-03), we retrieved 18 entries for FDA-approved compounds that have the term “helicase” as a target (Table 2). It is plausible that most of these molecules have already been tested during this emergency period caused by COVID-19, with unknown large-scale effect. In addition to these molecules, we found another two FDA-approved small compounds with experimental evidence of activity on viral helicases, guanidine hydrochloride (GuHCl) and ebselen.
GuHCl, a well-known small compound FDA-approved to treat the symptoms of muscle weakness and fatigability associated with Eaton-Lambert syndrome, was found able to inhibit activities of RNA helicase-like proteins, such as NS3 in Norovirus and VP35 in Ebola virus [27, 28]. However, this drug may present common side effects such as nausea, diarrhoea, stomach cramps, abnormal liver function tests and loss of appetite.
Ebselen is an additional molecule that may inhibit the RNA helicase NS3 of hepatitis C virus [29]. In this study, after a screening of 727 compounds in the National Institute of Health clinical collection sets and their testing for inhibitory activity on NS3 helicase binding nucleic acids, the authors found that ebselen was able to disrupt NS3/nucleic acid interactions and impair virus replication. Interestingly, ebselen is a selenium-containing compound that was tested in different Phase 1 and 2 clinical trials on patients with Meniere’s Disease or in subjects affected by type1/2 diabetes (https://www.drugbank.ca/drugs/DB12610). A double-blind, placebo-controlled, Phase 2 trial conducted in 83 healthy adults aged 18–31, demonstrated that ebselen treatment was well tolerated [30]. Interestingly, very recently, preliminary data demonstrated that ebselen strongly inhibits SARS-CoV-2 growth in a cell-based assay suggesting this as a molecule of particular interest for further investigation in clinical trials [31].
4 Future Directions and Conclusion
The disease caused by the new coronavirus is cause for an unprecedented concern for public health. As of April 28 2020, more than 200 countries have been affected by COVID-19, the total number of cases was 2,954,222 and deaths were approximately 202,600 worldwide. By comparison, SARS-CoV affected 26 countries and resulted in 8098 cases and 774 deaths from 2002 to 2004; and MERS-CoV affected 27 countries and caused 2519 cases and 858 deaths in 2012 (https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200428-sitrep-99-covid-19.pdf?sfvrsn=119fc381_2).
As of today, no FDA-approved therapies to treat COVID-19 or other coronaviruses exist. However, FDA updates every day guidelines for conducting clinical trials or diagnostic studies (https://www.fda.gov/emergency-preparedness-and-response/mcm-issues/coronavirus-disease-2019-covid-19). Moreover, as recently emerged from the work by Zhou et al. [8], the search and use of FDA-approved or investigational drugs that are outside the scope of the original clinical indication, currently known as drug repositioning, could be a promising approach to find a rapid strategy to limit the effects of the COVID-19 pandemic. In our opinion, the research on the G4s/helicase world could accelerate the finding of non-toxic effective therapeutics for SARS-CoV (Fig. 1), as well as for SARS-CoV-2, by the discovery of new G4 ligands rapidly transferable in the clinic, and by the employment of FDA-approved helicase inhibitors.
Moreover, a strategy that designs or repurposes drugs that exert their antiviral activity by inhibiting highly conserved mechanisms essential to viral replication, may reduce viral titres and improve host response in the event of future emergence of other human coronaviruses.
References
Zhong NS, Zheng BJ, Li Y, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–8.
Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92.
Stadler K, Masignani V, Eickmann M, et al. SARS–beginning to understand a new virus. Nat Rev Microbiol. 2003;1:209–18.
Ramadan N, Shaib H. Middle East respiratory syndrome coronavirus (MERS-CoV): a review. Germs. 2019;9:35–42.
Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–62.
Thorlund K, Dron L, Park J, et al. A real-time dashboard of clinical trials for COVID-19. Lancet Digital Health. 2020. https://doi.org/10.1016/S2589-7500(20)30086-8.
Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–8.
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14.
Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–37.
Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–515.
Fay MM, Lyons SM, Ivanov P. RNA G-quadruplexes in biology: principles and molecular mechanisms. J Mol Biol-. 2017;429:2127–47.
Métifiot M, Amrane S, Litvak S, Andreola ML. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res-. 2014;42:12352–66.
Ruggiero E, Richter SN. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018;46:3270–83.
Puig Lombardi E, Londoño-Vallejo A, Nicolas A. Relationship Between G-quadruplex sequence composition in viruses and their hosts. Molecules. 2019;24:E1942.
Lavezzo E, Berselli M, Frasson I, et al. G-quadruplex forming sequences in the genome of all known human viruses: a comprehensive guide. PLoS Comput Biol. 2018;14:e1006675.
Tan J, Vonrhein C, Smart OS, et al. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G quadruplexes. PLoS Pathog. 2009;5:e1000428.
Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–22.
Puig Lombardi E, Londoño-Vallejo A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020;48:1–15.
Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–82.
Mendoza O, Bourdoncle A, Boulé JB, Brosh RM Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006.
Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78:9977–86.
Briguglio I, Piras S, Corona P, Carta A. Inhibition of RNA helicases of ssRNA(+) virus belonging to flaviviridae, coronaviridae and picornaviridae families. Int J Med Chem. 2011;2011:213135.
Tanner JA, Zheng BJ, Zhou J, et al. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem Biol. 2005;12:303–11.
Cho JB, Lee JM, Ahn HC, Jeong YJ. Identification of a novel small molecule inhibitor against SARS coronavirus helicase. J Microbiol Biotechnol. 2015;25:2007–10.
Shum KT, Tanner JA. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. ChemBioChem. 2008;9:3037–45.
Zou X, Wu J, Gu J, Shen L, Mao L. Application of aptamers in virus detection and antiviral therapy. Front Microbiol. 2019;10:1462.
Li TF, Hosmillo M, Schwanke H, et al. Human norovirus NS3 Has RNA helicase and chaperoning activities. J Virol. 2018;92:e01606–17.
Shu T, Gan T, Bai P, et al. Ebola virus VP35 has novel NTPase and helicase-like activities. Nucleic Acids Res. 2019;47:5837–51.
Mukherjee S, Weiner WS, Schroeder CE, et al. Ebselen inhibits hepatitis C virus NS3 helicase binding to nucleic acid and prevents viral replication. ACS Chem Biol. 2014;9:2393–403.
Kil J, Lobarinas E, Spankovich C, et al. Safety and efficacy of ebselen for the prevention of noise-induced hearing loss: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;390:969–79.
Jin Z, Du X, Xu Y, et al. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020. https://doi.org/10.1038/s41586-020-2223-y(Epub ahead of print).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported by Bambino Gesù Children’s Hospital and Italy Ministry of Health.
Conflicts of interest
Nadia Panera, Alberto Tozzi and Anna Alisi, declare they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Panera, N., Tozzi, A.E. & Alisi, A. The G-Quadruplex/Helicase World as a Potential Antiviral Approach Against COVID-19. Drugs 80, 941–946 (2020). https://doi.org/10.1007/s40265-020-01321-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40265-020-01321-z