Abstract
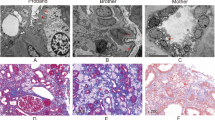
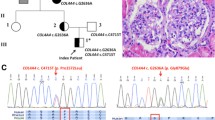
Alport syndrome (AS) is a rare hereditary disease that presents with chronic kidney disease and sensorineural hearing loss, and is diagnosed by its clinical features, pathological features on renal tissue, and mode of inheritance. We report a woman in her 20 s who exhibited persistent haematuria with normal renal function and sensorineural hearing loss. Her family members exhibited the same clinical findings among three generations and were suspected of having autosomal dominant AS (ADAS). Renal biopsy showed minor glomerular abnormalities on light microscopy and extensive thinning of the glomerular basement membrane on electron microscopy. Whole-exome analysis revealed a known COL4A4 (type IV collagen α4) mutation (c. 2510 G > C: p. Gly837Ala). Two pedigrees with the same variant have been reported previously, one as ADAS and the other as autosomal recessive AS. However, these two cases exhibited no sensorineural hearing loss. The analysis in the present case revealed another missense variant in ESPN (Espin), an actin-bundling protein, which is a causative gene for sensorineural hearing loss. Although the pathophysiological significance of this novel missense variant needs to be clarified, computational analysis predicted that the variant creates a new phosphorylation site for protein kinase C. Our case suggests a possible association of hereditary sensorineural hearing loss with ADAS. Whole-exome analysis should be considered to diagnose hereditary and multiple-organ disorders.


Similar content being viewed by others
Data availability
The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AS:
-
Alport syndrome
- ADAS:
-
Autosomal dominant Alport syndrome
- ARAS:
-
Autosomal recessive Alport syndrome
- XLAS:
-
X-linked Alport syndrome
References
Nozu K, Nakanishi K, Abe Y, Udagawa T, Okada S, Okamoto T, et al. A review of clinical characteristics and genetic backgrounds in Alport syndrome. Clin Exp Nephrol. 2019;23:158–68.
Kamiyoshi N, Nozu K, Fu XJ, Morisada N, Nozu Y, Ye MJ, et al. Genetic, clinical, and pathologic backgrounds of patients with autosomal dominant Alport syndrome. Clin J Am Soc Nephrol. 2016;11:1441–9.
Kremer H. Hereditary hearing loss; about the known and the unknown. Hear Res. 2019;376:58–68.
Donaudy F, Zheng L, Ficarella R, Ballana E, Carella M, Melchionda S, et al. Espin gene (ESPN) mutations associated with autosomal dominant hearing loss cause defects in microvillar elongation or organisation. J Med Genet. 2006;43:157–61.
Naz S, Gfiffith AJ, Riazuddin S, Hampton LL, Battey JF Jr, Khan SN, et al. Mutations of ESPN cause autosomal recessive deafness and vestibular dysfunction. J Med Genet. 2004;41:591–5.
Boulouiz R, Li Y, Soualhine H, Abidi O, Chaflk A, Nürnberg G, et al. A novel mutation in the Espin gene causes autosomal recessive nonsyndromic hearing loss but no apparent vestibular dysfunction in Moroccan family. Am J Med Genet A. 2008;23:3086–9.
Saida K, Kim CA, Ceroni JRM, Bertola DR, Honjo RS, Mitsuhashi S, et al. Hemorrhagic stroke and renovascular hypertension with Grange syndrome arising from a novel pathogenic variant in YY1AP1. J Hum Genet. 2019;64:885–90.
Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Oka M, Nozu K, Kaito H, Fu XJ, Nakanishi K, Hashimura Y, et al. Natural history of genetically proven autosomal recessive Alport syndrome. Pediatr Nephrol. 2014;29:1535–44.
Zheng L, Beeler DM, Bartles JR. Characterization and regulation an additional actin-filament-binding site in large isoforms of the stereocilia actin-bundling protein espin. J Cell Sci. 2014;127:1306–17.
Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49.
Huang HD, Lee TY, Tzeng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33:W226–W22929.
Acknowledgements
We thank the patient and her family for taking part in this study.
Funding
This research received no specific grant from any funding agency.
Author information
Authors and Affiliations
Contributions
YI designed and YI, YM, and YA organized the study. YI and TM cared for the patient. TN, MN, and YK performed renal biopsy and prepared the samples. YK provided pathological diagnoses. KS, NM, and NM performed whole-exome analysis and Sanger sequencing. YI and KS performed computational analysis. YI, AH, RM, YN, and MM reviewed the literature and interpreted the data obtained from all procedures. YI and MM wrote the manuscript, which was edited by all other authors. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no conflict of interest exists.
Ethics approval
All procedures performed in the studies were in accordance with the Ethical Committee of Kumamoto University Graduate School of Medical Sciences.
Consent for publication
The patients provided written informed consent to participate in this study. The patients provided written informed consent to publish this case report, including case description, medical data, and images, maintaining anonymity.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Izumi, Y., Hamaguchi, A., Miura, R. et al. Autosomal dominant Alport syndrome due to a COL4A4 mutation with an additional ESPN variant detected by whole-exome analysis. CEN Case Rep 9, 59–64 (2020). https://doi.org/10.1007/s13730-019-00429-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13730-019-00429-w