Abstract
Key message
High levels of genetic diversity, pronounced genetic structure and limitations to gene flow in Serbian spruce, a rare and endangered tree species from the refugial Balkan region, point towards a “one population-one unit” strategy for assembling a network of Genetic Conservation Units (GCUs) for its dynamic conservation. On the other hand, genetic information also permits to prioritize populations for conservation based on their contribution to genetic diversity and differentiation.
Context
Serbian spruce, Picea omorika (Panč.) Purk., is a rare, IUCN red-listed European conifer endemic to the Balkan region. Its current rigid conservation (without any intervention allowed in ~ 30 remnant populations) and the extant network of Genetic Conservation Units (four natural populations and three planted stands from the western part of the species range, in the Republic of Srpska, Bosnia and Herzegovina, RS-BH) might be ineffective in preserving the species’ genetic diversity.
Aims
To facilitate implementation of dynamic conservation of Serbian spruce by re-assessing the number and size of remnant populations in RS-BH and updating genetic knowledge on these understudied western Serbian spruce populations.
Methods
Comprehensive field survey in RS-BH, genotyping 689 individuals from 14 western populations with ten highly informative nuclear EST-SSRs and analytical methods for prioritizing populations for conservation based on their contribution to the geographical structuring of genetic diversity.
Results
The genetic diversity of western Serbian spruce populations (Ae = 2.524, HE = 0.451) is comparable with what was found for eastern ones; they are highly genetically differentiated (Hedrick’s G’ST = 0.186; Jost’s D = 0.097) and comprise ten distinct gene pools. Effective population size is often ≥ 15. As much as 14% of alleles is not preserved in the extant GCUs established in natural populations. Eight populations positively contribute to within-population genetic diversity, four to genetic differentiation, and two are globally important in terms of diversity and differentiation. Although wildfires may contribute to admixture of different gene pools, re-establishment from seeds from extirpated populations has likely prevailed in studied populations.
Conclusions
A larger network of GCUs is required for the dynamic conservation of western Serbian spruce populations. A “one population-one unit” strategy, with 14 GCUs, would represent the safest approach to conserve species extant genetic variation in this part of the species range. Nonetheless, a strategy to prioritize populations for conservation based on their contribution to allelic diversity has been put forward. Given the rapid global warming and peculiarities of Serbian spruce distribution, habitat and life history traits, conservation measures based on a rigorously designed GCU network are urgent for its rescue and survival.
Similar content being viewed by others
1 Introduction
The dynamic conservation of forest genetic resources (FGR), aimed at maintaining genetic diversity of forest tree species over time and preserving evolutionary processes and adaptive potential in their populations (Ledig 1986; Eriksson et al. 1993; Namkoong 1997), is considered a promising alternative to standard in situ and ex situ conservation strategies (Fady et al. 2016). In actively managed Genetic Conservation Units (GCUs), spontaneous mating and reproduction are allowed (Eriksson et al. 1993), thus providing conditions suitable for evolutionary forces to act on gene frequencies (Lefèvre et al. 2013). The existing network of GCUs (natural populations and/or field collections of individuals) covers different ecogeographical zones and is expected to capture the extant genetic diversity of a species. Therefore, GCUs comprising a network are carefully selected based on the conservation objective (large or marginal populations, endangered species) and genetic knowledge on natural population of the species in question (Koskela et al. 2013; Lefèvre et al. 2013).
The progress in the dynamic conservation of FGR has recently been assessed in 33 European countries (Lefèvre et al. 2013) on the basis of information available in the European Information System on Forest Genetic Resources (EUFGIS 2007–2011). This synthesis clearly showed the low representation of Mediterranean environmental zones in the FGR conservation network. However, the Mediterranean Basin represents a forest biodiversity hotspot (Médail and Quézel 1999; Fady-Welterlen 2005) with relict tree populations of a disproportionate importance for the conservation of European FGR and may comprise locally adapted populations potentially interesting for species’ adaptation to climate change (Petit et al. 2005). Therefore, Lefèvre et al. (2013) recommended increasing conservation efforts in this part of Europe, which is still genetically understudied (e.g. Hewitt 2000; Médail and Diadema 2009; Piotti et al. 2017).
Serbian spruce, Picea omorika (Panč.) Purk., occurs naturally within an area of c. 200 km2 in the central Balkans, around the mid-course of the Drina River, along the border between the Republic of Srpska, Bosnia and Herzegovina (RS-BH) and the Republic of Serbia (RS) (Aleksić and Geburek 2014; Ivetić and Aleksić 2016, 2019; Ballian et al. 2016). Due to the increased risk of extinction associated with such limited natural range and low number of remnant and small-sized populations susceptible to man-made and natural disturbances, this species is protected by the law in former Yugoslavia since 1955 and IUCN red-listed since 1998 (Conifer Specialist Group 1998). Without any available genetic knowledge on Serbian spruce, Yugoslav authorities assumed that the best way to ensure its long-term survival was to enable free development of natural populations, and thus applied rigid species-orientated conservation strategy without any intervention allowed. Later on, category 1 of the Protected Area Management Categories of IUCN and category 1.1 of the Ministerial Conference on the Protection of Forests in Europe (FOREST EUROPE-MCPFE) were allocated to all Serbian spruce populations, and this level of protection remained to the present days in both countries, RS-BH and RS, that were established at the end of the last century.
Serbian spruce was included into the pan-European network of GCUs (Lefèvre et al. 2013) with seven units (EUFGIS 2007–2011) designated following the list of seed stands in the RS-BH, i.e. in western part of the species range (the Register of seeds stands in the RS-BH, Mataruga et al. 2005). However, none of the units (four natural populations and three planted stands) fulfils the requirements given by Koskela et al. (2013) for designation of GCUs. That is, none of the GCUs is actively managed due to the rigid conservation strategy implemented in 1955. In addition, the utility of three planted stands which, in general, may be used as GCUs (Koskela et al. 2013), is questionable, because they were established from material of unknown origin (Mataruga et al. 2005). Furthermore, the genetic knowledge on the four natural populations, now GCUs, is rather limited because they have been studied to date only by Ballian et al. (2006). These authors analysed 12 western populations with allozymes and found low levels of genetic diversity and lack of genetic structure among studied populations. However, contrasting findings were obtained for more frequently studied eastern populations from RS, analysed with highly informative nuclear microsatellites (EST-SSRs) (Aleksić et al. 2009; Aleksić and Geburek 2010, 2014; Aleksić et al. 2017a). Each out of the ten eastern populations represents an independent, highly genetically diverse gene pool (Aleksić and Geburek 2014) and furthermore, a strong within-population genetic structure, with three spatially clustered gene pools, is present even in a small-sized population (418 trees in 0.35 ha) in this part of the species range distribution (Aleksić et al. 2017a).
Although the differences in genetic layouts between eastern and western populations might be partially explained by different environmental pressures in the two areas, e.g. wildfires which were more common in the western part of the species range over the past 100 years (Fukarek 1951a; Čolić 1987), the contrasting findings even between studies on western populations based on the same type of markers are also present (Kuittinen et al. 1991 vs. Ballian et al. 2006). According to Aleksić et al. (2009), the reasons for that are to be found in the usage of several loci in the study of Ballian et al. (2006) which cannot be characterized as polymorphic based on the frequency of detected alleles. These findings altogether indicate the need for further genetic surveys based on high-resolution molecular markers to assist development of correct dynamic conservation strategies. The drought-induced physiological weakening of Serbian spruce trees followed by infestation by Armillaria ostoyae (Romang.) Herink. and the rapid dieback of trees recently observed in Serbia (Ivetić and Aleksić 2016) are warnings showing the need for genetically informed conservation actions to maintain the evolutionary potential of this fragile (Ivetić and Aleksić 2016, 2019) and old spruce species (e.g. Lockwood et al. 2013; Chen et al. 2019). These activities are needed immediately considering the scarce natural regeneration recorded in almost all natural populations of this pioneer species (Čolić 1957; Aleksić and Geburek 2014) and lack of suitable new habitats at higher elevations within the species natural range for climate change-driven migrations (Ivetić and Aleksić 2016). In the meanwhile, it is essential to shed more light on the genetic layout of western populations because four of them, along with three planted stands, are the only GCUs currently available for Serbian spruce.
The aim of our study is to facilitate implementation of dynamic conservation of Serbian spruce by (i) re-assessing the number and size of remnant populations in the western part of the species range distribution, in RS-BH, and (ii) updating previous knowledge on the genetic layout of these Serbian spruce populations using high-resolution genetic data comparable to those available for eastern populations. For this purpose, we carried out a comprehensive field survey in RS-BH in order to record all extant Serbian spruce populations in this part of the species range distribution. We then analysed 689 individuals originating from 14 western natural populations by means of ten polymorphic EST-SSRs to elucidate the spatial distribution of genetic variation of Serbian spruce in RS-BH. Results were discussed to the light of available knowledge about the species’ life history traits and potential environmental stressors, in particular wildfires. Genetic data were finally used to provide guidelines for optimizing the dynamic conservation of this species in two steps. They comprise the improvement of current in situ conservation actions, which will enable short-term safeguarding of the species’ genetic diversity, until conditions for the second step, i.e. scientifically based long-term ex situ measures, are met.
2 Material and methods
2.1 Detection and census sizes of western Serbian spruce populations and sampling activities
All Serbian spruce natural populations in the RS-BH are located in public forests and are managed by the Forest Enterprise “Forests of the Republic of Srpska”. Following maps on distribution of Serbian spruce populations in RS-BH given by Plavšić (1936, 1937a, 1937b), Piškorić (1938), Fukarek (1951b), Ballian et al. (2006) and IUCN (Aleksić et al. 2017b) as well as suggestions of local foresters, all listed sites with Serbian spruce were visited during the summer 2016, and presence/absence of viable populations was recorded. The sizes of populations (in ha) were estimated from the ground, by calculating surface areas of occupancy limited by at least six trees found on population edges. Based on that, census sizes of populations, Nc, were estimated. At each site, the extent of both natural regeneration and dieback of individuals were recorded.
In total, 689 individuals from 14 populations were sampled, from 29 to 67 individuals per population (Table 1). Cambium samples were taken from 29 trees from population Stru and 19 trees from population Tiso because it was impossible to reach branches from the ground, while young needles were collected from the remaining trees. Samples were collected from adult individuals with dbh > 15 cm. Sampled trees were evenly distributed throughout populations and were selected at random along transects. Needles and cambium samples were dried at room temperature and kept at − 20 °C prior to DNA extraction.
2.2 Genotyping with nuclear microsatellites
Total genomic DNA was extracted from both needles and cambium samples following CTAB method of Doyle and Doyle (1990). Before extraction, plant material was lyophilized for 24 h and homogenized (ten needles per sample, 20 mg of cambium) with a swing mill (MM 400, Retsch GmbH, Germany). DNA extracts were fluorometrically quantified (Gene Quant Pro, Amersham Bioscience) and adjusted to 20 ng/μl.
Ten EST-SSRs were used for genotyping, eight published by Rungis et al. (2004) and two by Fluch et al. (2011). They were assembled into three multiplexes (MP) for parallel PCR amplification: MP1 (WS0022.B15, WS0092.M15 and WS00716.F13, annealing temperature Ta = 53 °C), MP2 (WS0073.H08 and WS00111.K13, Ta = 55 °C), MP3 (Pa_47, Pa_51, WS0032.M17, WS0046.M11 and WS0092.A19, Ta = 62 °C). Reverse primers from each primer pair were labeled with fluorescent dyes Cy5, IRD700 (both Biomers) and DY751 (Metabion).
Parallel PCR amplification of loci was performed with Type-it Microsatellite PCR kit (Qiagen, GmbH, Hilden, Germany) for each multiplex separately. PCR reactions were carried out in the total volume of 15 μl comprising 1 μl of 20 ng/μl genomic DNA, 7.5 μl of 2 × Type-it Multiplex PCR Master Mix, 5 μl of RNase free water and 1.5 μl of primer mix, respectively. PCR amplification was performed with a thermocycler (Biometra, Analytic Jena) following conditions: initial denaturation at 95 °C for 15 min; 30 amplification cycles of denaturation at 94 °C for 30 s, annealing according to multiplex for 90 s and extension at 72 °C for 30 s; final extension of 30 min at 60 °C. PCR amplification products were separated via capillary electrophoresis using 8-capillary DNA Analyzer automated sequencer [GenomeLab™ GeXP Genetic Analysis System (GeXP), AB Sciex]. Sizing of fragments was performed manually with Fragment Analyzer tool implemented in GenomeLab Software.
The occurrence of possible genotyping errors (stuttering, null alleles and large allele dropout) was tested using MICRO-CHECKER 2.2.1 (van Oosterhout et al. 2004) with 1000 randomizations.
2.3 Data analyses
Standard genetic diversity parameters (number of alleles, A; effective number of alleles, Ae; observed and expected heterozygosity, HO and HE, allelic richness, Ar; and private allelic richness, PAr) were calculated by GenAlEx 6.5 (Peakall and Smouse 2012) and hp-rare (Kalinowski 2005). Deviations of genotypic distributions from Hardy–Weinberg expectations (HWE) and genotypic linkage disequilibrium (LD) between all pairs of loci were assessed using GENEPOP 4.0 (Rousset 2008). The effective population size (Ne) was assessed based on the linkage disequilibrium method implemented in NeEstimator v2 (Do et al. 2014), discarding alleles with frequencies < 0.01.
Parameters of genetic differentiation among populations (G’ST, Hedrick 2005, and Jost’s D, Jost 2008) were estimated with GenAlEx 6.5, and their statistical significance was assessed with 999 permutations. The same software was used for assessing the effective number of migrants per generation (Nm) calculated as 1/4[(1/FST)-1)] and for principal coordinate analysis (PCoA) on a matrix of pairwise genetic distances between populations (Peakall and Smouse 2012). The analysis of molecular variance (AMOVA, Excoffier et al. 1992) was carried out with Arlequin ver. 3.5 (Excoffier et al. 2005). The distance method was the number of different alleles, and the significance of tests was examined by 15, 000 permutations.
A measure of the contribution of each population to the total allelic diversity (AT) and its within-subpopulation (AS) and between-subpopulation (DA) components was estimated with metapop2 (López-Cortegano et al. 2019).
The Bayesian clustering algorithm implemented in STRUCTURE v. 2.3 (Pritchard et al. 2000) was used to assess the putative number of differentiated genetic clusters. The most likely number of clusters (K) in which individuals can be divided was assessed using the empirical statistic ΔK calculated by STRUCTURE HARVESTER (Evanno et al. 2005; Earl and von Holdt 2012). The default parameter settings of the admixture model with correlated allele frequencies and no LOCPRIOR were used, with K varying from 1 to 15. Each run consisted of 1 × 105 burn-in iterations and 5 × 105 data collection iterations, and ten runs for each K were performed. Different runs for the same K were averaged using the software CLUMPP (Jakobsson and Rosenberg 2007). To further investigate the possible presence of a genetic structure among clusters found at the strongest level of structuring (i.e. K = 2, see Results), STRUCTURE was also run within each genetic cluster on individuals assigned with q > 0.80 with the same settings as described above.
The heterozygosity excess test in BOTTLENECK 1.2.02 (Piry et al. 1999) was used to infer possible occurrence of a recent bottleneck in the last 2 Ne–4 Ne generations. The two-phase-mutation model was used, with variance of 30% and the proportion of stepwise mutations of 70%. Statistical significance was assessed using the one-tailed Wilcoxon test.
The isolation-by-distance (IBD) pattern (Wright 1943) was tested by performing a Mantel test (Mantel 1967) of pairwise population’s genetic distance matrices [G’ST/(1-G’ST)] on the linear geographic distance matrix, using GenAlEx with 999 permutations.
3 Results
3.1 Detection and census sizes of western Serbian spruce populations
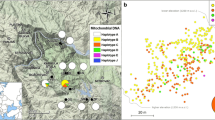
We recorded 14 viable Serbian spruce populations (Nc ≥ 50) in the RS-BH (Table 1, Fig. 1). Two formerly large populations, Pliština and Božurevac, are nowadays represented by < 10 individuals. The former was destroyed by wildfire in 1990–1995; the latter is severely suppressed by the advancement of more competitive species. Therefore, they were not considered as populations but as groups of trees and were not sampled. One additional viable population was recorded but not sampled because it was surrounded by mine fields (Novo Brdo). It is worth noting that three additional populations cited in the literature (i.e. Plavšić 1936, 1937a, 1937b; Piškorić 1938; Fukarek 1951b) were not found. They should be considered as eradicated unless the information about their locations was not erroneous.
Distribution of remnant Serbian spruce populations within an area of c. 200 km2. The main part of the species distribution range is around the mid-course of the Drina River, in area west of the river (eastern part of the Republic of Srpska, Bosnia and Herzegovina, RS-BH) and east of the river (western part of the Republic of Serbia, RS) in the central Balkans. Locations of three disjunct populations are shown in the right lower figure. All western populations from the RS-BH were used in the present study (black circles). Eastern populations from the RS (white circles, labelled with capital letters) were studied elsewhere
Estimated areas occupied by populations ranged from 0.4 (Vidi) to 60.8 ha (Vel_St) and Nc from 50 (Panj) to 10,000 individuals (Vel_St) (Table 1). Poor natural regeneration was observed at all sites, and only sporadic dieback of individuals was recorded, mostly at low altitude, in all populations.
3.2 Genotyping and genetic diversity parameters
Ten microsatellite loci were successfully amplified in 689 individuals. The percentage of missing data was 0.46%, and most of them were in Stru (4.75% at ten loci in 40 individuals), probably due to the lower quality of DNA from cambium samples. The possible presence of null alleles was low and inconsistent among populations. Null alleles were only recorded at locus WS00111.K13 in two populations (Stru and Sta_st), and at locus WS00716.F13 in one population (Vel_St), while errors in genotyping due to the stuttering and allele dropout were not present.
The total number of alleles at ten loci in 689 individuals was 82. Ae and Ar58 in overall sample were 2.524 and 4.098, respectively (Table 1). Five populations harboured 17 private alleles, and average PAr58 was 0.110. Overall, HO and HE were 0.465 and 0.451, respectively. The distribution of Ar58 and PAr58 in populations is shown in Fig. 2. Eight populations contributed positively to total allelic diversity (Fig. 3). Among these, three populations (Stru, Vel_St and Vijo) were the largest contributors to within-population allelic diversity. A small but significant excess of heterozygotes was observed in six populations and in overall sample. After Bonferroni adjustments of significance levels for multiple tests, significant genotypic LD was detected in 98 out of 630 tests, with ten significant tests recorded in population Stru. Ne ranged from 6.4 [3.3, 11.3] in Panj to ∞ [49.2, ∞] in Per_ga.
Severe size reductions in the last 2 Ne–4 Ne generations were observed in six populations of which four were located in the north-west part of the species range in RS-BH (Grad, Panj, Sar_bu and Tiso), and two in the central part of the range (Suv_do and Kar_st) (Appendix Table 4). According to available literature on effects of wildfires on Serbian spruce populations over the past 100 years (full or partial devastation of populations, Fukarek 1951a; Čolić 1987), all these populations were re-established at sites severely devastated by wildfire either 70 years ago (Grad, Sar_bu, Suv_do and Kar_st) or more recently (Panj) with the exception of population Tiso. Recent bottlenecks were not observed in populations Sta_st and Per_ga established at sites burned over the past 100 years, nor in those less affected by past wildfires (Gost, Rado, Stru, Vel_St and Vijo).
3.3 Genetic structure of populations
The lowest pairwise G’ST and D values were 0.028 and 0.017, respectively, between Vel_St and Kar_st, and the highest values were 0.271 and 0.185, respectively, between Panj and Vijo (Table 2 and Appendix Table 5). All values were statistically significant (P < 0.001). G’ST and D values in overall sample were 0.186 (± 0.044) and 0.097 (± 0.040), respectively. Genetic structuring of populations was indicated also by AMOVA. The 10.15% of molecular variation was assigned to variation among populations (Table 3). The detected amount of genetic differentiation among populations was mainly due to the contribution of the two disjunct populations, Vijo and Rado, plus the population with both the highest allelic richness and the largest contribution to total allelic diversity (Stru) (Fig. 3).
Bayesian clustering supported the presence of a strong genetic structure within western Serbian spruce natural populations (Fig. 4), with optimal grouping at K = 2 (according to Evanno method) and K = 10 (according to the likelihood profile, Appendix Figure 7). Clustering of populations at K = 2 approximately followed a north-south direction (Appendix Figure 8). Six neighbouring northern populations (Grad, Panj, Sar_bu, Sta_st, Stru and Tiso) plus Gost and the disjunct Rado comprised the first gene pool, while the remaining nearby central populations (Kar_st, Per_ga, Suv_do, Vidi and Vel_St) plus the disjunct Vijo formed the second one. At K = 10, more than 50% of individuals from eight populations were strongly assigned (q ≥ 0.80) to their own distinct gene pool, while less than 50% of individuals from the two nearby populations Grad and Sar_bu, 3 km apart, were strongly assigned to the ninth gene pool (the remaining individuals were strongly assigned to neighbouring gene pools). The same holds for another pair of populations, Panj and Sta_st (9.71 km apart), while Vel_St and Kar_st, less than 1 km distant, comprised individuals strongly assigned mainly to the gene pool of their closest neighbour, Vidi, and also to gene pools specific for neighbouring populations.
In order to test whether delineation of ten distinct gene pools is justified, we carried out two additional STRUCTURE analyses with both individuals strongly assigned to the “blue” and “red” gene pools shown in Fig. 4. The first analysis included 294 individuals originating from all populations except Vijo, which lacked trees strongly assigned to the “blue” gene pool, and the optimal number of genetic groups was two (according to Evanno method) to six (according to the likelihood profile, Appendix 5a). The second analysis with 264 individuals from all populations except Panj and Rado, which lacked trees strongly assigned to the “red” gene pool, revealed that the optimal number of gene pools was three to seven (Appendix 5b).
PCoA mostly confirmed the genetic structure highlighted by Bayesian clustering, showing the main genetic relationships among populations (Fig. 5). The first two principal coordinates explained 29.94% and 15.15% of variation in genetic distances, respectively. The first PCo separated populations into two main groups mostly resembling STRUCTURE results at K = 2. The position of populations along the second PCo is concordant with what was indicated by Bayesian clustering at K = 10. As an example, three nearby central populations Vidi, Vel_St and Kar_st clustered together and separate from the three southern populations Per_ga, Suv_do and Gost. The two disjunct populations, Vijo and Rado, are at the margins of the PCoA scatterplot confirming the large number of cluster detected by STRUCTURE analyses both on the entire dataset and on the main genetic clusters detected at the uppermost hierarchical level of the genetic structure.
Outcomes of the principle coordinates analysis (PCoA). The scores of populations are given in the space defined by the first and second coordinate and are presented as pies showing the assignment probability of each population to genetic clusters inferred at K = 10 in Bayesian clustering shown in Fig. 4
The number of migrants per generation, reflecting historical gene flow, ranged from 1.11 between Panj and Vijo (distant 39.25 km) to 8.95 migrants per generation between Kar_st and Vel_St (distant 0.72 km), with average number of migrants per generation of 2.68 in overall sample (average distance between populations 23.26 km) (Appendix Table 6). Upon exclusion of two disjunct populations, overall Nm increased to 3.09 migrants per generation between populations distant 11.94 km on average. A significant correlation between geographic and genetic distance for all pairwise populations was found (r = 0.613, P < 0.001, Fig. 6). Such IBD patter was characterized by a steep relationship up to 20 km, when a plateau was reached.
4 Discussion
The vulnerability of Serbian spruce due to its limited natural range and the low number of small-sized remnant populations makes it a priority target for conservation actions (Ivetić and Aleksić 2016, 2019). The onset of conservation initiatives in Serbian spruce dates back to 1955 (Ivetić and Aleksić 2016, 2019). This species was IUCN-red listed later on (Conifer Specialist Group 1998), and a network of seven GCUs, aimed at maintaining the species’ adaptive potential (Koskela et al. 2013; Lefèvre et al. 2013), has been recently established (EUFGIS 2007–2011). However, we demonstrate that ongoing conservation measures, not based on genetic knowledge, are likely to be ineffective in achieving their goals. Given the already visible detrimental effects of the global warming on Serbian spruce (e.g. the drought-induced physiological weakening of trees followed by infestation by Armillaria ostoyae observed in RS, Ivetić and Aleksić 2016), its exceptionally limited dispersal ability (Aleksić and Geburek 2014; Aleksić et al. 2017a ), and the lack of suitable habitats at higher elevations within the species’ natural range, Serbian spruce persistence in its natural habitat in the next decades is uncertain unless effective conservation measures are undertaken (Ivetić and Aleksić 2016, 2019). Our work, along with previous genetic studies on Serbian spruce (e.g. Aleksić and Geburek 2010, 2014), represents the necessary starting point to improve existing in situ conservation efforts and to set up ex situ conservation actions. These actions should be coupled with the establishment of an efficient GCU network for the dynamic conservation of this exceptional and fragile forest genetic resource.
4.1 Current census data of western Serbian spruce populations
Although the natural range of Serbian spruce is nowadays only ~ 200 km2, the exact number and sizes of its remnant fragmented populations are uncertain, and this represents the first obstacle for any conservation action (Frankham et al. 2002). For instance, an accurate distribution map of Serbian spruce is still lacking, and the most comprehensive one available to date is that provided within the most recent IUCN report (Aleksić et al. 2017b), which is concordant with the first descriptions of the species’ natural range (e.g. Plavšić 1936, 1937a, 1937b; Piškorić 1938; Fukarek 1951b). However, the most recent literature about the species genetic layout accounts for often contrasting descriptions of extant populations (e.g. Ballian et al. 2006 and Nasri et al. 2008 vs. Aleksić et al. 2017b). The inconsistency about the number and sizes of remnant Serbian spruce populations most likely results from the lack of monitoring of demographic changes of populations in both RS-BH and RS since the first descriptions of the species distribution (Plavšić 1936, 1937a, 1937b; Piškorić 1938; Fukarek 1951b). In the meanwhile, frequent wildfires affected ~ 40% of populations, especially in the western part of its range (Fukarek 1951a; Čolić 1987). Although some populations were completely devastated more than once, most of them successfully re-established at burned sites (Čolić 1987).
After a comprehensive field survey carried out during the summer 2016, we recorded 15 viable Serbian spruce populations with Nc ≥ 50 in the western part of the species distribution, of which 14 were included in the present study. Although these estimates have to be taken with caution because we used rather unsophisticated methods for their assessment, they represent a first step towards clarifying the current size of western Serbian spruce populations which might be useful until more precise data will be collected. The future usage of up-to-date methods for assessing the size of forest patches, such as remote sensing based on LIDAR technology, will most likely provide improved estimates of areas occupied by populations. However, considering that Serbian spruce often vegetates in terrains characterized by extreme steepness and roughness (Ballian et al. 2016) and/or in mixed forests at highly heterogeneous density (Aleksić and Geburek 2010), we deem that the estimates of census size presented here, carried out by observers with > 10-year experience in working with Serbian spruce populations, will represent an important benchmark for comparing data collected by interpretation of aerial images.
4.2 Genetic layout of western Serbian spruce populations
The second obstacle for the development of conservation actions for Serbian spruce is the insufficient and often contrasting genetic information on its populations. To overcome this obstacle, we provided high-resolution genetic data for genetically understudied western populations, comparable to those available for eastern populations, and demonstrate the similarity of the main features of the genetic structure of western and eastern populations.
Western populations are characterized by slightly lower levels of genetic diversity (HE ranging from 0.397 to 0.497) as well as effective population sizes (Ne < 50 in ten populations) than eastern ones (Aleksić and Geburek 2014). The number of migrants per generation together with the IBD pattern detected showed a steep relationship between geographic and genetic distance up to 20 km. Average and pair-wise differentiation indices are markedly high considering what was usually found for other conifers with similar dispersal syndromes at larger spatial scales (e.g. Tollefsrud et al. 2009; Awad et al. 2014; Wagner et al. 2015; Piotti et al. 2017). These findings are consistent with previous indications pointing towards limitations to gene flow in Serbian spruce (Aleksić and Geburek 2014; Aleksić et al. 2017a) despite its rather light pollen and seeds (Kohlermann 1950). Such limited but still effective genetic connectivity likely determined the presence of ten distinct gene pools among the 14 populations included in our study.
Another phenomenon accounting for the observed genetic structure among western populations is the source-sink dynamics determined by wildfires (Shohami and Nathan 2013). As already mentioned, wildfires were more frequent in the western part of the species range distribution over the past 100 years (Fukarek 1951a; Čolić 1987), and the most devastating ones, occurring during 1946–1947, were most likely deliberately caused by local people. Available data on effects of wildfires on Serbian spruce populations demonstrate that viable populations were re-established at more or less all sites burned 70 years ago (Čolić 1987). This is in accordance with findings on the light requirements for the development of Serbian spruce seedlings (Tucić and Stojković 2001) which suggest facilitated natural regeneration of this pioneer species at open sites as opposed to dense forests (when density can reach 1200 ind/ha, Aleksić et al. 2017a). Re-establishment of Serbian spruce following disturbances such as wildfires was also common during the Holocene history of this species (Finsinger et al. 2017). These pieces of evidence would imply that wildfires might play an important role not only for natural regeneration of the species but also in modelling the genetic structure of its populations. However, genetic consequences of wildfire on Serbian spruce have not been studied to date. Although the quality of available data on wildfires does not allow a quantitative assessment of their influence on the distribution of genetic diversity in Serbian spruce, the comparison of results between populations differently affected by wildfires allowed us to make some preliminary considerations about this issue.
We found that almost all western populations affected by wildfire during the 20th century harbour signs of the recent bottleneck, and interestingly, they usually keep their genetic distinctiveness suggesting prevalent development of seedlings from locally surviving trees. For instance, in an area where five adjacent populations were present before the wildfire in 1946–1947 (Fukarek 1951b; Čolić 1987), two populations are currently found, Grad and Sar_bu. They are distant 3 km, and mainly comprise individuals strongly assigned (q ≥ 0.80) to the same and unique gene pool. Another pair of populations, characterized by similar disturbance history and a unique shared and predominant gene pool, are Panj and Sta_st, 10 km distant. However, in STRUCTURE analyses at K ≥ 10, they were split into different gene pools, each corresponding to one population (data not shown). This would suggest that the effects of genetic drift can grow faster at a geographic distance higher than 3 km, where gene flow is likely extremely difficult (Aleksić and Geburek 2014; Aleksić et al. 2017a). On the other hand, a population fully destroyed and recovered two times over the past 100 years, Kar_st, is highly genetically similar to the nearby population Vel_St (0.72 km distant), exposed to multiple but less severe wildfires in the past (Čolić 1987). Although such considerations are based on limited and outdated literature on the occurrence of wildfire in Serbian spruce populations (e.g. Fukarek 1951b; Čolić 1987), they point towards the importance of nearby seed sources for the species to be able to recover after severe disturbances.
4.3 Implications for the dynamic conservation of Serbian spruce
The current network of Serbian spruce GCUs comprises seven units in the western part of the species range (EUFGIS 2007–2011). The size of three GCUs (BIH00001, BIH00094, BIH00093) corresponds roughly to half of the extant size of natural populations Stru, Tiso and Vel_St, respectively. The area of the fourth GCU (BIH00069) is almost four times larger than the actual size of the Gost population (25.8 ha vs. 7.1 ha), which is not expected in case of the species-orientated conservation strategy with a single target species (Koskela et al. 2013; Lefèvre et al. 2013). Disregarding issues related to the size of extant GCUs covering western natural populations, we found that as much as 14% of alleles of western populations are not present in the extant network of four GCUs. The percentage of uncaptured alleles would most likely increase substantially if eastern, genetically distinct and highly diverse populations (Aleksić and Geburek 2014) are also taken into consideration.
The remaining three GCUs (Table 1) were not included into our study because they are planted stands established with the material of unknown origin (Mataruga et al. 2005) which cannot be resolved with low-resolution allozyme markers (Ballian 2006). Since it is unreasonable to allocate resources on conservation of unknown and potentially depleted material, keeping planted stands in the GCU network for Serbian spruce is not recommended at this stage because of the lack of knowledge on their conservation values as GCUs. Although we do not recommend their removal, as it might be useful when plantations represent a relevant source of pollen to small autochthonous stands (Santini et al. 2018), they may be re-purposed. They have been used to date for studying, for instance, seed germination (Cvjetković et al. 2013). Altogether, this would imply that the current network of GCUs in Serbian spruce is highly inefficient in maintaining available genetic diversity in this species, and thus, it should be improved.
Considering the genetic structure emerging from our analyses, i.e. ten highly diverse but poorly connected and differentiated gene pools, and the facts that almost all studied populations have defined surface areas and Ne ≥ 15, thus fulfilling the basic requirements for designation as GCUs (Koskela et al. 2013), the most parsimonious strategy to select populations for the dynamic conservation of western Serbian spruce FGR should be to delineate at least ten GCUs. In this case, eight GCUs would correspond to distinct populations, and two would encompass only one population from each of the two pairs of genetically distinct but admixed populations belonging to the same gene pool (Grad/Sar_bu, and Panj/Sta_st).
However, since the management of a large number of GCUs requires substantial labour and financial resources, an available option is to prioritize populations for conservation based on their contribution to within- and between-population allelic diversity (López-Cortegano et al. 2019). Overall, nine populations positively contribute to single or both components of allelic diversity, and the effort needed for their conservation is in line with what emerged from the analyses of the genetic structure. Among analysed populations, a triplet showed an outstanding contribution to allelic diversity. Population Stru, from the core of the species range, and the disjunct population Vijo are globally important because of their high and positive total contribution for both allelic diversity and differentiation. The second studied disjunct population, Rado, stands out for its substantial positive contribution to the between-population component of allelic diversity. Although such ranking of populations relative to their contribution to total allelic diversity and its components facilitates delineation of the most important ones for conservation purposes, it is worth noting that focusing financial resources and conservation action only on three populations might be a risky strategy in an environment particularly prone to wildfires (Fukarek 1951a; Čolić 1987) and for an endangered species (Aleksić et al. 2017b).
On the other hand, in case that the labour and financial aspects would not be an issue, the safest strategy to conserve Serbian spruce extant genetic variation is to designate all populations that fulfil criteria recommended by Koskela et al. (2013) as GCUs. This is reasonable because Serbian spruce populations are expected to follow different evolutionary trajectories in terms of the rather limited gene flow typical for this species (Aleksić and Geburek 2014; Aleksić et al. 2017a), and because conservation value of certain admixed populations may not be clearly evident based on nuclear data. For instance, only western populations Kar_st and Vel_St harbour, at low frequency, mitochondrial haplotypes (D. Gömöry, unpublished data) exclusively found in a single eastern population (Aleksić and Geburek 2014; Aleksić et al. 2017a). Since two populations included in our study (i.e. Gost and Vel_St) were also analysed by Aleksić and Geburek (2014), it is likely that at least 22 GCUs are needed to preserve most of the genetic variation currently available in the natural range of the species. However, such consideration at the species level should be taken with caution because a study on all Serbian spruce natural populations relaying on the same marker set is not available yet.
Our suggestion to include all Serbian spruce populations into the network of GCUs points towards a “one population-one unit” strategy for delineation of GCUs which has not been proposed previously for any rare and endangered tree species in Europe and worldwide (e.g. EUFGIS 2007–2011; Li et al. 2018). Nonetheless, such strategy is well-supported by comprehensive genetic surveys (our study and the study of Aleksić and Geburek 2014) that highlight a specific genetic structure in Serbian spruce (high within- and among-population diversity, poor connectivity of populations, and Ne ≥ 15) which is not concordant with expectations on genetic layouts of tree species with limited natural range and fragmented distribution of small-sized populations (Leimu et al. 2006; Honnay and Jacquemyn 2007). The observed genetic structure in Serbian spruce results from the species long-term persistence in a glacial refugium in which maintenance of high levels of genetic diversity and high genetic differentiation of populations were enabled through the specific population dynamics over time (Aleksić and Geburek 2014). Thus, our approach may be suitable for populations of tree species confined to glacial refugia.
Since active intervention is not allowed in Serbian spruce natural populations due to the current rigid conservation strategy applied in RS-BH and RS, it is necessary to change relevant legislation frameworks in both countries, i.e. to allocate category 1.2 of the FOREST EUROPE-MCPFE to designated GCUs, in order to allow management with active intervention. Selective removal of competitors will facilitate otherwise poor natural regeneration in populations, while prescribed burning programmes aimed at reducing species vulnerability (Finsinger et al. 2017) might be recommended only when aimed at reducing competition to promote the expansion of extant populations. On the other hand, considering the peculiar genetic structure of the species, prescribed burning programmes potentially affecting extant populations should be strictly avoided. Planting of seedlings and/or direct seeding from carefully selected sources is expected to increase the resilience of populations and to ensure their persistence and recovery after a catastrophe (Koskela et al. 2013). We have to emphasize that these actions, which require substantial resources, may not succeed in enabling even the short-term survival of Serbian spruce (Ivetić and Aleksić 2016, 2019). This is because of the detrimental effects of the climate warming on this cold-adapted species and rather slim odds to survive in situ even in the near future (Ivetić and Aleksić 2016, 2019). However, they are necessary until conditions for implementation of ex situ measures are met. These measures, which comprise establishment of seed banks, seed orchards, clone collections as well as a controversial approach-assisted migration, require planning, commitment, resources and coordinated actions of scientists and relevant authorities in two countries, RS-BH and RS.
5 Conclusions
\Current efforts on Serbian spruce conservation, implemented in the mid-20th century without any genetic knowledge on the species, will be likely ineffective in maintaining the species genetic diversity and preserving its adaptive potential. We showed that the extant GCU network does not adequately cover the genetic diversity of the species. Given the peculiar genetic structure likely linked to both historical factors and gene flow limitations (Aleksić and Geburek 2014; Aleksić et al. 2017a), we argue that selecting a limited number of GCUs for the species’ dynamic conservation might be risky. Nonetheless, a prioritized list of populations for conservation has been provided based on the genetic information collected. Such ranking can be useful in case of need to optimize the allocation of scarce resources, but should be taken with caution considering the fire-prone environment hosting Serbian spruce. We also stress that resources allocated to the in situ conservation of Serbian spruce will enable only its short-term survival because of the potentially detrimental effect of climate warming on this cold-adapted species. Nonetheless, these measures are essential to safeguard sufficient genetic diversity for the implementation of scientifically based ex situ measures.
Abbreviations
- A :
-
number of alleles
- Ar 58 :
-
allelic richness
- A S :
-
within-population allelic diversity
- A T :
-
total allelic diversity
- BH:
-
Bosnia and Herzegovina
- D :
-
Jost’s genetic distance
- D A :
-
between-population allelic diversity
- EST-SSR:
-
expressed-sequence-tagged simple sequence repeat
- EUFGIS:
-
European Information System on Forest Genetic Resources, www.eufgis.org
- FGR:
-
forest genetic resources
- F IS :
-
fixation index
- GCU:
-
genetic conservation unit
- G’ ST :
-
Hedrick’s genetic distance
- H O :
-
observed heterozygosity
- H E :
-
expected heterozygosity
- IUCN:
-
International Union for the Conservation of Nature
- MCPFE:
-
Ministerial Conference on the Protection of Forests in Europe
- Na :
-
effective number of alleles
- Ne :
-
effective population size
- PA :
-
private alleles
- PAr 58 :
-
private allelic richness
- RS:
-
Republic of Serbia
- RS-BH:
-
Republic of Srpska, Bosnia and Herzegovina
References
Aleksić JM, Geburek T (2010) Mitochondrial DNA reveals complex genetic structuring in a stenoendemic conifer Picea omorika [(Panč.) Purk.] caused by its long persistence within the refugial Balkan region. Plant Syst Evol 285:1–11. https://doi.org/10.1007/s00606-009-0250-0
Aleksić JM, Geburek T (2014) Quaternary population dynamics of an endemic conifer, Picea omorika, and their conservation implications. Conserv Genet 15:87–107. https://doi.org/10.1007/s10592-013-0523-6
Aleksić JM, Schueler S, Mengl M, Geburek T (2009) EST-SSRs developed for other Picea species amplify in Picea omorika and reveal high genetic variation in two natural populations. Belg J Bot 142:89–95 https://www.jstor.org/stable/20794675
Aleksić JM, Piotti A, Geburek T, Vendramin GG (2017a) Exploring and conserving a “microcosm”: whole-population genetic characterization within a refugial area of the endemic, relict conifer Picea omorika. Conserv Genet 18:777–788. https://doi.org/10.1007/s10592-017-0926-x
Aleksić JM, Ballian D, Isajev D, Mataruga M, Christian T, Gardner M (2017b) Picea omorika. The IUCN Red List of Threatened Species 2017:e.T30313A84039544. https://doi.org/10.2305/IUCN.UK.2017-2.RLTS.T30313A84039544.en.
Aleksić JM, Piotti A, Geburek T, Vendramin GG, Mataruga M, Danicic V, Cvjetkovic B, Fussi B, Konnert M (2019) Microsatellite data for 689 Picea omorika (Serbian spruce) trees from 14 western populations from the Republic of Srpska, Bosnia and Herzegovina. Version 15 June 2019. FigShare [Dataset]. https://doi.org/10.6084/m9.figshare.8280767.v1
Awad L, Fady B, Khater C et al (2014) Genetic structure and diversity of the endangered fir tree of Lebanon (Abies cilicica Carr.): implications for conservation. PLoS One 9:e90086. https://doi.org/10.1371/journal.pone.0090086
Ballian D (2006) Controlling the Serbian spruce origin (Picea omorika Panč./Purk.) at the Kakanj plantation with isoenzyme markers. Šumarski list 7-8:295–304 (in Croatian with English summary)
Ballian D, Longauer R, Mikić T, Paule L, Kajba D, Gömöry D (2006) Genetic structure of a rare European conifer, Serbian spruce (Picea omorika (Pančić) Purk.). Plant Syst Evol 260:53–63. https://doi.org/10.1007/s00606-006-0431-z
Ballian D, Ravazzi C, Caudullo G (2016) Picea omorika in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A (eds) European Atlas of Forest Tree Species. Publications Office of the European Union, Luxembourg, pp 117–117
Chen J, Li L, Milesi P, Jansson G, Berlin M, Karlsson B, Aleksic J, Vendramin GG, Lascoux M (2019) Genomic data provides new insights on the demographic history and the extent of recent material transfers in Norway spruce. Evol Appl. https://doi.org/10.1111/eva.12801
Čolić D (1957) Neke pionirske karakteristike omorike (Picea omorika Panč.) i njihova uloga u sukcesiji biljnih zajednica. Arch Biol Sci 9:51–60 (in Serbian with English summary)
Čolić D (1987) Spontana obnova Pančićeve omorike (Picea omorika Panč.) posle požara. Zaštita Prirode 40:37–56 (in Serbian with English summary)
Conifer Specialist Group (1998) Picea omorika. 2007 IUCN Red List of Threatened Species. http://www.iucnredlist.org/search/details.php/30313/summ
Cvjetković B, Mataruga M, Isajev V, Lević J, Lučić A, Trkulja V, Kremenović Ž (2013) Variability in germination and germination dynamics of differently treated seeds of Serbian spruce (Picea omorika Pančić/Purkynĕ). Genetika 45:109–119
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214. https://doi.org/10.1111/1755-0998.12157
Doyle JJ, Doyle LJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Eriksson G, Namkoong G, Roberds JH (1993) Dynamic gene conservation for uncertain futures. For Ecol Manag 62:15–37. https://doi.org/10.1016/0378-1127(93)90039-P
EUFGIS (2007-2011) Establishment of a European Information System on Forest Genetic Resources. www.eufgis.org. Accessed 7 November 2018
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50. https://doi.org/10.1177/117693430500100003
Fady B, Aravanopoulos FA, Alizoti P, Mátyás C, von Wühlisch G, Westergren M et al (2016) Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. For Ecol Manag 375:66–75. https://doi.org/10.1016/j.foreco.2016.05.015
Fady-Welterlen B (2005) Is there really more biodiversity in Mediterranean forest ecosystems? Taxon 54:905–910
Finsinger W, Morales-Molino C, Gałka M, Valsecchi V, Bojovic S, Tinner W (2017) Holocene vegetation and fire dynamics at Crveni Potok, a small mire in the Dinaric Alps (Tara National Park, Serbia). Quat Sci Rev 167:63–77. https://doi.org/10.1016/j.quascirev.2017.04.032
Fluch S, Burg A, Kopecky D, Homolka A, Spiess N, Vendramin GG (2011) Characterization of variable EST SSR markers for Norway spruce (Picea abies L.). BMC Res Notes 4:401. https://doi.org/10.1186/1756-0500-4-401
Frankham R, Ballou J, Briscoe D (2002) Introduction to Conservation Genetics, 1st edn. Cambridge University Press, Cambridge
Fukarek P (1951a) Staništa Pančićeve omorike nakon šumskih požara 1946/47. godine. Šumarski list 75:61–71 (in Serbian with English summary)
Fukarek P (1951b) Današnje rasprostranjenje Pančićeve omorike (Picea omorika Pančić) i neki podaci o njenim sastojinama. Godišnjak Biološkog Instituta u Sarajevu III/1–2:141–198 (in Serbian with German summary)
Hedrick PW (2005) A standardized genetic differentiation measure. Evol Int J Org Evol 59:1633–1638. https://doi.org/10.1111/j.0014-3820.2005.tb01814.x
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907. https://doi.org/10.1038/35016000
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831. https://doi.org/10.1111/j.1523-1739.2006.00646.x
Ivetić V, Aleksić JM (2016) Response of rare and endangered species Picea omorika to climate change: The need for speed. Reforesta 1:81–89. https://doi.org/10.21750/REFOR.2.09.24
Ivetić V, Aleksić JM (2019) Serbian spruce and climate change: Possible outcomes and conservation strategy. In: Šijačić-Nikolić M, Milivanović J, Nonić M (eds) Forests of Southeast Europe Under a Changing Climate. Advances in Global Change Research, vol 65. Springer, Cham, pp 353–371
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026. https://doi.org/10.1111/j.1365-294X.2008.03887.x
Kalinowski ST (2005) hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Kohlermann L (1950) Untersuchungen über die Windverbreitung der Früchte und Samen mitteleuropäischer Waldbäume. Forstwiss Cbl 69:606–624
Koskela J, Lefèvre F, Schueler S, Kraigher H, Olrik DC, Hubert J et al (2013) Translating conservation genetics into management: Pan-European minimum requirements for dynamic conservation units of forest tree genetic diversity. Biol Conserv 157:39–49. https://doi.org/10.1016/j.biocon.2012.07.023
Kuittinen H, Muona O, Karkkainen K, Borzan Ž (1991) Serbian spruce, a narrow endemic, contains much genetic variation. Can J For Resour 21:363–367. https://doi.org/10.1139/x91-044
Ledig FT (1986) Conservation strategies for forest gene resources. For Ecol Manag 14:77–90
Lefèvre F, Koskela J, Hubert J, Kraigher H, Longauer R, Olrik DC et al (2013) Dynamic conservation of forest genetic resources in 33 European countries. Conserv Biol 27:373–384. https://doi.org/10.1111/j.1523-1739.2012.01961.x
Leimu R, Mutikainen P, Koricheva J, Fisher M (2006) How general are positive relationships between plant population size, fitness and genetic variation? J Ecol 94:942–952. https://doi.org/10.1111/j.1365-2745.2006.01150.x
Li S, Gan X, Han H, Zhang X, Tian Z (2018) Low within-population genetic diversity and high genetic differentiation among populations of the endangered plant Tetracentron sinense Oliver revealed by inter-simple sequence repeat analysis. Ann For Sci 75:74. https://doi.org/10.1007/s13595-018-0752-4
Lockwood JD, Aleksić MJ, Zou J, Wang J, Liu J, Renner SS (2013) A new phylogeny for the genus Picea from plastid, mitochondrial and nuclear sequences. Mol Phylogenet Evol 69:717–727. https://doi.org/10.1016/j.ympev.2013.07.004
López-Cortegano E, Pérez-Figueroa A, Caballero A (2019) metapop2: Re-implementation of software for the analysis and management of subdivided populations using gene and allelic diversity. Mol Ecol Resour. https://doi.org/10.1111/1755-0998.13015
Mantel N (1967) The detection of disease clustering and generalize regression approach. Cancer Res 27:209–220
Mataruga M, Isajev V, Lazarev V, Balotić P, Daničić V (2005) Registar šumskih sjemenskih objekata RS. Osnova unapređenja sjemenske proivodnje. Šumarski fakultet Univerziteta u Banjoj Luci, Art print. (in Serbian)
Médail F, Diadema K (2009) Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr 36:1333–1345. https://doi.org/10.1111/j.1365-2699.2008.02051.x
Médail F, Quézel P (1999) Biodiversity hotspots in the Mediterranean Basin: setting global conservation priorities. Conserv Biol 13:1510–1513. https://doi.org/10.1046/j.1523-1739.1999.98467.x
Namkoong G (1997) A gene conservation plan for loblolly pine. Can J For Res 27:433–437. https://doi.org/10.1139/x96-196
Nasri N, Bojović S, Vendramin GG, Fady B (2008) Population genetic structure of the relict Serbian spruce, Picea omorika, inferred from plastid DNA. Plant Syst Evol 271:1–7. https://doi.org/10.1007/s00606-007-0594-2
Peakall R, Smouse PE (2012) GenAlEx 6.5, genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28:2537–2539 http://bioinformatics.oxfordjournals.org/content/28/19/2537
Petit RJ, Hampe A, Cheddadi R (2005) Climate changes and tree phylogeography in the Mediterranean. Taxon 54:877–885
Piotti A, Leonarduzzi C, Postolache D, Bagnoli F, Spanu I, Brousseau L, Urbinati C, Leonardi S, Vendramin GG (2017) Unexpected scenarios from Mediterranean refugial areas: disentangling complex demographic dynamics along the Apennine distribution of silver fir. J Biogeogr 44:1547–1558. https://doi.org/10.1111/jbi.13011
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Piškorić O (1938) Prilog poznavanju omorike (Picea omorika Pančić). Šumarski list 62:577–585 (in Serbian)
Plavšić S (1936) Staništa Pančićeve omorike na levoj obali Drine (s kartom u tekstu). Glasnik zemaljskog muzeja u Bosni i Hercegovini, God. XLVIII, druga sveska, Sarajevo: 17–26. (in Bosnian)
Plavšić S (1937a) Istorija otkrića i proučavanja Pančićeve omorike. Glasnik zemaljskog muzeja u Bosni i Hercegovini, God. XLIX, druga sveska, Sarajevo:35–48. (in Bosnian)
Plavšić S (1937b) Staništa Pančićeve omorike u okolini Foče (s kartom u tekstu). Glasnik zemaljskog muzeja u Bosni i Hercegovini, God. XLIX, druga sveska, Sarajevo:29–34. (in Bosnian)
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106. https://doi.org/10.1111/j.1471-8286.2007.01931.x
Rungis D, Bérubé Y, Zhang J, Ralph S, Ritland CE, Ellis BE, Douglas C, Bohlmann J, Ritland K (2004) Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theor Appl Genet 109:1283–1294. https://doi.org/10.1007/s00122-004-1742-5
Santini F, Andrisano T, Leonardi S, Ciaschetti G, Labriola M, Vendramin GG, Piotti A (2018) Tracking the origin of silver fir plantations along the boundary between different genetic clusters in central Apennines: Implications for their management. For Ecol Manag 408:220–227. https://doi.org/10.1016/j.foreco.2017.10.045
Shohami D, Nathan R (2013) Fire-induced population reduction and landscape opening increases gene flow via pollen in Pinus halepensis. Mol Ecol 23:70–81. https://doi.org/10.1111/mec.12506
Tollefsrud MM, Sønstebø JH, Brochmann C et al (2009) Combined analysis of nuclear and mitochondrial markers provide new insight into the genetic structure of North European Picea abies. Heredity 102:549–562. https://doi.org/10.1038/hdy.2009.16
Tucić B, Stojković B (2001) Shade avoidance syndrome in Picea omorika seedlings: a growth-room experiment. J Evol Biol 14:444–455. https://doi.org/10.1046/j.1420-9101.2001.00291.x
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Wagner S, Liepelt S, Gerber S, Petit RJ (2015) Within-range translocations and their consequences in European larch. PLoS One 10:e0127516. https://doi.org/10.1371/journal.pone.0127516
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgements
The authors would like to express their gratitude to forest engineers from the Forest Enterprise “Šume Republike Srpske” a.d. Sokolac for help in the field, colleagues from ASP-Bayerisches Amt für forstliche Saat-und Pflanzenzucht, Teisendorf, Germany for technical support, and dr. M. Marchi, PhD, for graphical presentation of the Serbian spruce distribution in Republic of Srpska, Bosnia and Herzegovina.
Statement on data availability
Microsatellite data are freely available on Figshare (Aleksić et al. 2019) at https://doi.org/10.6084/m9.figshare.8280767.v1
Funding
This work was supported by the Ministry of Science and Technology of the Republic of Srpska Government, grant number 19/6-020/961-50, ASP-Bayerisches Amt für forstliche Saat-und Pflanzenzucht, Teisendorf, Germany, and Forest Enterprise “Šume Republike Srpske” a.d. Sokolac.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Bruno Fady
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1
Appendix 2
Appendix 3
Inferring optimal number of genetic clusters from STRUCTURE analysis based on (a) distribution of Pr(K) at K ranging from 1 to 15 (vertical bars represent standard errors) (left) and (b) ΔK method of Evanno et al. (2005) (right)
Appendix 4
Outcomes of the Bayesian clustering presented according to the assignment probability of each population to genetic clusters inferred at K = 2 (left) and K = 10 (right). Outcomes of the Bayesian clustering based on individual q values at K = 2 and K = 10 are given in Fig. 4
Appendix 5

Appendix 6
Rights and permissions
About this article
Cite this article
Mataruga, M., Piotti, A., Daničić, V. et al. Towards the dynamic conservation of Serbian spruce (Picea omorika) western populations. Annals of Forest Science 77, 1 (2020). https://doi.org/10.1007/s13595-019-0892-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-019-0892-1