Abstract
Acne vulgaris is the most common dermatological disorder worldwide. It is a multifactorial disease that involves increased sebum production, hyperkeratinization of the pilosebaceous unit, Propionibacterium acnes (Cutibacterium acnes) colonization, and inflammation. The human skin microbiome hosts a wide variety of microorganisms, including bacteria, viruses, and fungi. A delicate balance of these microorganisms is essential for the barrier function of the skin. Propionibacterium acnes represents nearly 90% of the human skin microbiome of healthy adults. Acne is a chronic recurrent disease that requires long-lasting treatment, which has led to the emergence of antibiotic resistance. New alternatives to traditional therapy are emerging, including antimicrobial peptides, natural engineered antibodies, and bacteriophages. Bacteriophages have been shown to play a role in human skin health and disease. There is evidence supporting phage therapy in many types of skin infections. P. acnes bacteriophages have been isolated and characterized. However, only a few in vitro studies have tested the ability of bacteriophages to kill P. acnes. Furthermore, there is no evidence on bacteriophage therapy in the treatment of acne in humans. In this review, we summarize the most recent evidence regarding P. acnes bacteriophages and the potential role of these bacteriophages in the treatment of acne. Further research on this field will provide the evidence to use phage therapy to decrease rates of antibiotic resistance and restore antibiotic susceptibility of P. acnes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acne vulgaris is the most common dermatological disorder worldwide. It affects around 50 million people each year in the USA, with an estimated annual cost of $2.5 billion [1]. The worldwide prevalence of acne is estimated to be around 9% [2], accounting for 0.3% of the global disease burden [3]. Although acne affects people of all ages, 85% of all affected individuals are 12–24 years old [4,5,6,7]. Severe acne also carries a high social and psychological impact, affecting emotions, self-esteem, and increasing the risk of depression and suicide [7].
The role of Propionibacterium acnes in the pathophysiology of acne is still under debate. P. acnes is the predominant commensal microorganism of the human skin microbiome. A delicate balance within the skin microbiota is essential for the barrier function of the skin and prevention of pathogen colonization [8].
The emergence of antibiotic resistance has become a public health problem worldwide [9]. The long-term use of topical and systemic antibiotics has led to high rates of antibiotic-resistant P. acnes strains [10]. As research on new antibiotic agents is decreasing due to cost and difficulty, the development of new, natural, and non-conventional alternatives—such as antimicrobial peptides, natural engineered antibodies, and bacteriophages—is becoming critical. Bacteriophage therapy seems to be a promising alternative. Its advantages are host specificity and simplicity of isolation and production. Although both in vitro and in vivo studies have shown the potential of targeted bacteriophage therapy in skin infections, research is lacking on bacteriophage therapy targeting P. acnes-associated infections. It has recently been proposed that the species P. acnes be reclassified to Cutibacterium acnes and other genera [11]. Here, we use the old nomenclature (P. acnes) throughout because it is still used by most of the evidence presented in this review. In this review, we summarize the most recent evidence on P. acnes bacteriophages and its potential role in the treatment of acne. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Acne Vulgaris Pathophysiology
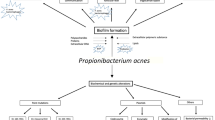
Acne is a multifactorial disease. Increased sebum production by androgen stimulation, abnormal hyperkeratinization of the pilosebaceous duct, and subsequent bacterial colonization and inflammation all contribute to the disease [12, 13]. It is proposed that P. acnes colonization plays a pivotal role in the pathogenesis of acne since antimicrobial therapy has been effective in treating acne for many years. However, its contribution to acne development is controversial [14].
Propionibacterium acnes is a Gram-positive, anaerobic/microaerophilic, fat-splitting, rod-shaped bacterium found on the skin; it represents nearly 90% of the skin microbiome of healthy adults [1, 14, 15]. The concentration of P. acnes depends on the abundance of sebaceous follicles and the age of the individual [13, 16, 17]. Accordingly, its concentration is higher on sebaceous areas such as the face, scalp, and back [13, 18], and various studies have reported an association between P. acnes levels and sebum production [16]. There is a marked increase in P. acnes colonization during puberty [15], which correlates with the time when sebaceous glands mature [13]. P. acnes may disrupt keratinocyte differentiation in the follicle, thereby contributing to the formation of comedones and inflammatory acne lesions by triggering a host inflammatory response [12, 19]. P. acnes produces enzymes that degrade skin components as well as chemotactic factors that stimulate keratinocytes and inflammatory cells to release pro-inflammatory cytokines (e.g., interleukin [IL]-8, IL-12, IL-1α, IL1-β, tumor necrosis factor alpha) and reactive oxygen species [1, 13, 20,21,22,23]. Although P. acnes is a commensal organism in humans, not all healthy adolescents or adults develop acne, indicating that differences in the pathogenicity of P. acnes strains must exist [1]. It has been proposed that certain strains play a pathogenic role and others act as bystanders [24,25,26,27]. Studies have also shown that specific genes in the P. acnes genome contribute to bacterium virulence and hence to acne pathophysiology [14, 26, 28].
Emerging Antibiotic Resistance
Over 2 million Americans become infected every year with antibiotic-resistant bacteria, resulting in about 23,000 deaths [29, 30]. The post-antibiotic era is approaching as antibiotic effectiveness steadily declines, and multiple common infections become resistant to treatment [14, 31]. The excessive use of antibiotics in agriculture and humans, the evolutionary pressure inherent to antibiotics [31], and the lack of research on new antibiotic agents are some of the reasons behind antibiotic resistance [14, 29, 31, 32].
Antibiotic resistance is among the main causes of treatment failure in acne vulgaris [33]. The mechanism of antibiotic resistance to P. acnes is explained by the remarkable genetic plasticity of bacteria [34]. Two major genetic strategies permit antimicrobial resistance: (1) gene mutation, and (2) foreign DNA coding acquisition through horizontal gene transfer [34]. Gene mutation is the predominant mechanism leading to P. acnes antibiotic resistance [12, 35].
Acne therapy warrants long-term treatment with topical and systemic antibiotics, which contributes to resistant P. acnes strains. Globally, P. acnes resistance to antimicrobials has increased almost 40% between the 1980s and 2000s worldwide [10]. Erythromycin/clindamycin-resistant P. acnes seems to be the most common pattern of resistance based on reports from the USA, Europe, and Asia [36,37,38]. However, resistance rates vary by region. Europe, Singapore, and Hong-Kong have high prevalence rates of erythromycin/clindamycin-resistant P. acnes (45–91%) and tetracycline-resistant P. acnes (2–26%) [36, 37, 39,40,41,42], but countries that practice conservative use of antibiotics, such as Japan and Korea, report much lower resistance rates (2–4%) [36, 42, 43].
The long-term use of antibiotics can promote the formation of an antibiotic-resistant biofilm that protects the bacterium against host defenses and can alter the natural microbiota of the skin [12, 44]. Studies have shown colonization by antibiotic-resistant coagulase-negative Staphylococci and Streptococcus pyogenes in acne patients who have used both topical and oral antibiotic therapy [45, 46]. Thereby, it is recommended to limit monotherapy with topical antibiotics and instead to combine them with other topical agents such as retinoids or benzoyl peroxide to decrease the risk of resistance [47].
Human Skin Microbiome
The human skin microbiome is home to a variety of microorganisms, including bacteria, fungi, viruses, and arthropods. A delicate balance between the microorganisms is essential for local immunity and barrier function of the skin [8]. Imbalances in this system have been linked to dermatologic diseases, such as acne, atopic dermatitis, psoriasis, and rosacea [48].
The dominant bacterial species found on adult skin are Propionibacterium, Corynebacterium, and Staphylococcus [48]. The skin can be divided into dry, moist, or sebaceous microenvironments, and each of these microclimates host varying proportions of these common bacterial flora [48].
Age-related shifts in bacterial communities could explain why certain skin diseases are prevalent at different stages of life. For example, the microbiome in children, who are more susceptible to atopic dermatitis, is composed of mostly Streptococcaceae, Bacteroides, and Proteobacteria [49]. Pubertal spikes in androgens lead to a more susceptible environment for the development of acne and a shift in microbiome concentration to greater levels of Propionibacterium and Corynebacterium.
Bacteriophages
Delving deeper into the bacteria of the microbiome brings us to bacteriophages. Bacteriophages, or viruses that infect bacteria, can be found throughout the biosphere, are essential members of the human microbiome, and may play an important regulatory role in human skin health and disease [8, 50,51,52]. Little is known about bacteriophage interaction with skin microbiota. Bacteriophages are obligatory intracellular parasites; thereby, their distribution depends on their host organisms [50]. There are over 6000 well-known bacteriophages [53], and these bacteriophages are estimated to be at least tenfold more common than bacteria [14]. There is wide diversity in the structure of these phages (e.g., tailed, polyhedral, pleomorphic, filamentous), and they are usually classified based on their genetic content [53].
Four distinct life cycle phases have been described: lytic (virulent phages), lysogenic, pseudo-lysogenic (temperate phages), and chronic infection [53] (Table 1). The first step in every phage cycle is the binding of the phage to bacterial surface receptors, after which the phage injects its genetic material (DNA or RNA) into the cells [54]. Phages undergoing the lytic phase, also called virulent phages, are the most abundant type and the most widely used in bacteriophage therapy due to their natural ability to kill bacteria directly through cell lysis.
Phages (temperate or dormant phages) that enter the lysogenic phase have the ability to induce transduction [53], a vital process in bacterial pathogenesis and adaptation [29, 55]. Transduction can confer advantages to bacteria by transferring pathogenicity or antibiotic resistance genes [14, 53, 56, 57]. Due to this risk, only those bacteriophages with lytic activity should be considered in phage therapy [53]. However, transduction can be used to our benefit by genetically engineering phages to transfer genes to reverse antibiotic resistance or to increase bacterial susceptibility to antibiotics [29]. Edgar et al. restored antibiotic sensitivity to streptomycin and nalidixic acid in resistant Escherichia coli in an in vitro study using genetically engineered bacteriophages [58]. In another study, Lue and Collins used genetically engineered phages to transfer genes that target DNA repair mechanisms, resulting in an increase in E. coli susceptibility to quinolones in vitro and in vivo [59]. These studies show that engineered bacteriophages can enhance the killing of antibiotic-resistant bacteria and biofilm and reduce the emergence of antibiotic resistance in bacteria.
Propionibacterium acnes Bacteriophages
Propionibacterium acnes bacteriophages were first identified by Brzin in 1964 [60]. In 1968, Zierdtc et al. isolated phage 174 from Corynebacterium acnes strains and used it to classify the Corynebacterium family, ultimately finding that 88% of all C. acnes strains were sensitive to this bacteriophage [54, 61]. A later study reported that 18% of P. acnes phages carried bacteriophages [62], and P. acnes bacteriophages were subsequently used to classify Corynebacterium and Propionibacterium [63, 64].
P. acnes bacteriophages are relatively more abundant in lipid-rich areas of the skin, correlating with the distribution of P. acnes in the skin [65,66,67]. These bacteriophages are the dominant phages in the pilosebaceous unit. Fitz-Gibbon et al. reported a 1:120 bacteriophages:P. acnes ratio in pilosebaceous units in healthy skin samples [26, 68, 69]. Most P. acnes bacteriophages possess a siphoviral morphology (i.e., isometric head and long flexible tail) [8, 15, 70, 71] and have a pseudolysogenic life cycle (Table 1) [8, 15, 72, 73]. P. bacteriophages displaying a lytic life cycle have also been characterized [70, 74].
Interestingly, despite the isolation of P. acnes bacteriophages over a varied temporal and geographical range, their genome is preserved with very limited genetic diversity [8, 15, 71, 72]. Marinelli et al. investigated the diversity of bacteriophages that infect P. acnes and isolated 11 P. acnes bacteriophages which lacked the genetic diversity seen in other phage populations [15]. A recent study by Liu et al. sequenced 48 P. acnes bacteriophages from human skin follicles and found a sequence identity of between 85 and 100% between strains, suggesting that the P. acnes bacteriophage population in the skin microbiota is dominated by one strain [8]. The authors tested the P. acnes–bacteriophage interaction and found that the 74 P. acnes strains were susceptible to the 15 tested P. acnes bacteriophages. They suggested multiple reasons for the lack of phage diversity, including a bottle-neck hypothesis leaving one dominant genotype, or the evolutionary constrains imposed on phages and bacteria to maintain a single phage, thus limiting the spreading of phage resistance [8]. Another possible explanation for the limited diversity of P. acnes bacteriophages could be due to the niche in which they live, as P. acnes makes up 90% of the microbiota of the pilosebaceous unit, thereby limiting horizontal gene transfer and increased diversity between phages in the pilosebaceous unit [14].
Liu et al. also found that some individuals shared the same bacteriophage strains in the skin microbiota, suggesting the existence of a pool of common bacteriophages among human populations [8]. They further discovered identical bacteriophages strains between closely related individuals (siblings), which makes human to human virus transmission a possibility. As seen in other studies [15], the resistance of certain P. acnes strains to bacteriophages was an issue [8]. Two possible resistance mechanisms are described by these authors, namely, restriction modification and clustered regularly interspaced short palindromic repeat (CRISPR), both of which target viral DNA integration into the host genome [8]. The findings of this study led the authors to conclude that the ability of P. acnes bacteriophages to lyse only susceptible strains may alter the bacteria population, as different strains will grow at different rates, thereby modulating the composition and dynamics of the skin microbiota.
The apparent lack of genetic diversity of P. acnes bacteriophages and their broad host range make them ideal candidates for phage therapy in acne [14]. Moreover, lytic bacteriophages engineered to target P. acnes strains in the specific microbiome of individuals will increase the success rate of acne treatments.
Propionibacterium acnes Bacteriophage Therapy
The potential role of bacteriophage therapy in acne vulgaris has recently attracted the interest of researchers and clinicians. Phages active against P. acnes have been isolated from the skin, oral cavity, and gastrointestinal tract [14]. It is important to note that only phages with proven lytic activity should be used in phage therapy because lysogenic or temperate phages carry the risk for transduction of antibiotic resistance or pathogenicity genes and may lead to delayed cell lysis [14, 53].
Bacteriophage therapy has been used in humans for several types of infections with good results [75,76,77,78]. However, no trials on P. acnes bacteriophage therapy have been conducted in humans. Brown et al. isolated ten bacteriophages capable of lysing P. acnes from human skin microbiota and tested their therapeutic potential [72]. These authors created a suspension for each bacteriophage at a final concentration of 2.5 × 108 PFU/g using an aqueous cetomacrogol cream that showed that these bacteriophage formulations effectively lysed P. acnes cells in agar lawn culture plates and remained active in the cream for up to 90 days when stored at 4 °C in light-protected bottles. The bacteriophage was specific to P. acnes strains and did not lyse other bacteria of the Propionibacterium family. Cells that regrew from the areas within the P. acnes plaques showed phage resistance [72]. Although some authors have suggested that a cocktail of phages could be used to decrease the risk of phage resistance [8, 15], P. acnes bacteriophage variability is relatively low, which may limit this approach. This important limitation needs to be explored in further studies. However, the results using the cream formulation of Brown et al. [72] suggest that P. acnes bacteriophage therapy is a simple and realistic therapeutic option for the treatment of acne. Another in vitro study showed effective eradication of P. acnes strains when P. acnes bacteriophages were isolated from human skin microbiota and applied in drops onto agar plates [79]. In this study, P. acnes bacteriophages were unable to kill other bacteria, such as Staphylococcus aureus, S. epidermidis, and Corynebacterium xerosis, confirming the specificity of these bacteriophages [79]. Formulations such as oil–base cream, water–oil nanoemulsion, biodegradable polyester matrix, antiseptic gel, and paraffin-oil-based lotion, have proven to be effective strategies to deliver bacteriophages [80].
P. acnes bacteriophage genomes encode endolysins involved in bacteria cell-wall degradation (muramidases, amidases, endopeptidases, glucosaminidases, and transglycosylases) [15, 70, 81]. These endolysins are implicated in the release of progeny following phage assembly by targeting peptidoglycan in the bacterial wall [70]. Marinelli et al. suggested that endolysins are a potential therapeutic option in acne therapy [15]. Phage endolysins are highly conserved in different P. acnes bacteriophage strains (95% at the amino acid level) [15], which implies that endolysins from any P. acnes bacteriophage could be active against most P. acnes strains. Phage endolysins have been used as antimicrobials both in vitro and in vivo with promising results [82]. Furthermore, no resistance to phage endolysins has been reported [82]. It has been shown that even bacteria that become phage resistant may remain endolysin sensitive [14]. This introduces yet another way to treat acne through the genetic engineering of enzymes to target bacteria cell walls.
These possibilities carry important therapeutic implications in the management of acne. Antibiotics, in combination with bacteriophage cocktails, could be used to decrease antibiotic resistance and to treat antibiotic-resistant P. acnes. However, further research is needed to evaluate P. acnes bacteriophage therapy in human subjects, both as monotherapy and in combination with conventional therapies.
Advantages of Phage Therapy
Bacteriophages have a low environmental impact compared to chemical antibiotics due to their natural origin [53, 83]. They target both Gram-positive and Gram-negative bacteria [84,85,86,87,88,89,90,91,92], and many in vitro and in vivo models have shown that bacteriophages are effective against multidrug-resistant bacteria [85,86,87,88]. As antibiotic resistance grows, phages retain the ability to kill antibiotic-resistant bacteria due to their differing mechanisms of action [53]. Bacteriophages are specific to their bacterial hosts (species), and only replicate locally, limiting the pressure on normal non-targeted flora of the skin and other organs [75, 93, 94]. Bacteriophages have also been shown to distribute in good concentrations all over the body, including the central nervous system [53, 91, 95] (Table 2).
Another potential benefit of bacteriophage therapy is the ability to decrease biofilm formation [83, 96,97,98,99,100,101]. Many in vitro and in vivo studies have proven that the combination therapy of antibiotics and lytic bacteriophages displays synergism by improving the efficacy of bacteria and biofilm eradication and preventing the emergence of resistant bacteria [92, 102,103,104,105,106,107,108,109,110,111]. Antibiotics could be conjugated with bacteriophages to deliver antibiotics to specific bacteria and at higher concentrations [14]. Furthermore, engineered bacteriophages could be used to improve efficacy through the transfer of susceptibility or sensitizing genes by means of genetic engineering.
The identification of bacteria and bacteriophage isolation for therapeutic purposes is a rapid and affordable process compared to the development of new antibiotics [93]. Moreover, the cost of bacteriophage therapy seems to be lower than that of traditional antibiotic therapy [112]; however, more studies are needed to establish the real short- or long-term costs of bacteriophage therapy.
Finally, bacteriophages are safe and well-tolerated, and no significant adverse events have been reported [75, 77, 78, 94, 113, 114].
Limitations of Bacteriophage Therapy
Although the prospect of using bacteriophage therapy to treat acne in a world with increasing antibiotic resistance is promising, this novel therapeutic endeavor comes with limitations. The first of these is our evolving understanding of phages and their life cycles. The newest data suggests that phages exist on a continuum between lytic and lysogenic life cycles [115]. This creates challenges when using phages as therapeutic vehicles since conventional phage therapy requires phages to undergo lytic cycles and rapidly kill their hosts. Most P. acnes phages characterized thus far, however, display pseudolysogeny (Table 1).
In addition, CRISPR protects bacteria from viral DNA integration, and P. acnes may become resistant to phage therapy through this mechanism [54]. This issue of resistance becomes even more likely with the knowledge that bacteriophages targeting P. acnes are highly homogeneous [26]. Therefore, the acquisition of resistance to one phage may confer P. acnes with resistance to many of its bacteriophages [14]. However, the risk is low compared to antibiotics, partially because bacteriophages can mutate and bypass bacteriophage resistance mechanisms [93]. Resistance can also be prevented by using multiple bacteriophages (cocktails) or synergistic combinations of bacteriophages + antibiotics [93].
In addition to these limitations, the more practical aspects of establishing optimal therapeutic doses, treatment frequency, and duration have not been established [54]. A more long-term risk with this therapy includes the unknown consequences to the cutaneous microbiome if P. acnes, a vital member to this community, is temporarily eradicated through acne treatment. We do not fully understand the repercussions of altering the natural micro-ecosystem of the skin.
Future therapeutic option
As the antibiotic resistance era approaches, research on new alternative antimicrobial agents is becoming critical. Bacteriophages, ubiquitous microorganisms of the human skin microbiome, contain distinct advantages that mark them as promising alternatives to conventional antimicrobial therapy. Among these advantages are host specificity, limited cross resistance, ease of isolation, low cost, and a favorable safety profile compared to antibiotics.
The potential clinical application of bacteriophage therapy for acne vulgaris is promising. Limitations to phage therapy, such as the risk of transduction of pathogenicity genes and the low P. acnes bacteriophage variability, can be overcome by a more thorough understanding of the bacteria–bacteriophage interaction in the human skin microbiome. Phage resistance is another limitation that must be considered and warrants further study.
As the field develops, more data is needed before phage therapy in human subjects is introduced. Developing targeted phage therapy, engineered bacteriophages, and enzyme-based therapies, either alone or as an adjuvant to antibiotics, may lead to decreasing rates of P. acnes resistance to antibiotics and the restoration of antibiotic susceptibility to P. acnes.
References
Zaenglein A, Thiboutot D. Acne vulgaris. In: Bolognia J, Schaffer J, Cerroni L, eds. Dermatology. 4th edn. Amsterdam: Elsevier; 2012:588–603.
Hay R, Johns N, Williams H, et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Investig Dermatol. 2013;134:1527–34.
Karimkhani C, Dellavalle RP, Coffeng LE, et al. Global skin disease morbidity and mortality: an update from the global burden of disease study 2013. JAMA Dermatol. 2017;153(5):406–12.
Lynn DD, Umari T, Dunnick CA, Dellavalle RP. The epidemiology of acne vulgaris in late adolescence. Adolesc Health Med Ther. 2016;7:13–25.
Sidbury R, Paller AS. The diagnosis and management of acne. Pediatr Ann. 2000;29(1):17–24.
White GM. Recent findings in the epidemiologic evidence, classification, and subtypes of acne vulgaris. J Am Acad Dermatol. 1998;39:S34–7.
Halvorsen JA, Stern RS, Dalgard F, Thoresen M, Bjertness E, Lien L. Suicidal ideation, mental health problems, and social impairment are increased in adolescents with acne: a population-based study. J Investig Dermatol. 2011;131(2):363–70.
Liu J, Yan R, Zhong Q, et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015;9(9):2078–93.
World Health Organization (WHO). Antimicrobial resistance: global report on surveillance. 2014. Available online at: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf. Accessed 30 June 2018.
Coates P, Vyakrnam S, Eady EA, Jones CE, Cove JH, Cunliffe WJ. Prevalence of antibiotic-resistant propionibacteria on the skin of acne patients: 10-year surveillance data and snapshot distribution study. Br J Dermatol. 2002;146(5):840–8.
Scholz CF, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66(11):4422–32.
Dessinioti C, Katsambas A. Propionibacterium acnes and antimicrobial resistance in acne. Clin Dermatol. 2017;35(2):163–7.
Liu PF, Hsieh YD, Lin YC, Two A, Shu CW, Huang CM. Propionibacterium acnes in the pathogenesis and immunotherapy of acne vulgaris. Curr Drug Metab. 2015;16(4):245–54.
Jonczyk-Matysiak E, Weber-Dabrowska B, Zaczek M, et al. Prospects of phage application in the treatment of acne caused by Propionibacterium acnes. Front Microbiol. 2017;8:164.
Marinelli LJ, Fitz-Gibbon S, Hayes C, et al. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. MBio. 2012;3(5):e00279. https://doi.org/10.1128/mBio.00279-12
McGinley KJ, Webster GF, Ruggieri MR, Leyden JJ. Regional variations in density of cutaneous propionibacteria: correlation of Propionibacterium acnes populations with sebaceous secretion. J Clin Microbiol. 1980;12(5):672–5.
Brzuszkiewicz E, Weiner J, Wollherr A, et al. Comparative genomics and transcriptomics of Propionibacterium acnes. PLoS One. 2011;6(6):e21581.
Kong HH, Segre JA. Skin microbiome: looking back to move forward. J Investig Dermatol. 2012;132(3 Pt 2):933–9.
Tyner H, Patel R. Propionibacterium acnes biofilm—a sanctuary for Staphylococcus aureus? Anaerobe. 2016;40:63–7.
Kim J, Ochoa MT, Krutzik SR, et al. Activation of toll-like receptor 2 in acne triggers inflammatory cytokine responses. J Immunol. 2002;169(3):1535–41.
Jeremy AH, Holland DB, Roberts SG, Thomson KF, Cunliffe WJ. Inflammatory events are involved in acne lesion initiation. J Investig Dermatol. 2003;121(1):20–7.
Vowels BR, Yang S, Leyden JJ. Induction of proinflammatory cytokines by a soluble factor of Propionibacterium acnes: implications for chronic inflammatory acne. Infect Immun. 1995;63(8):3158–65.
Nagy I, Pivarcsi A, Kis K, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 2006;8(8):2195–205.
Kwon H, Suh D. Recent progress in the research about Propionibacterium acnes strain diversity and acne: pathogen or bystander? Int J Dermatol. 2016;55(11):1196–204.
Jahns A, Lundskog B, Dahlberg I, Tamayo N, McDowell A, Patrick S, et al. No link between rosacea and Propionibacterium acnes. APMIS. 2012;120(11):922–5.
Fitz-Gibbon S, Tomida S, Chiu BH, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Investig Dermatol. 2013;133(9):2152–60.
Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One. 2010;5(8):e12277.
Lodes MJ, Secrist H, Benson DR, et al. Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology. 2006;152(Pt 12):3667–81.
Jassim A, Limoges R. Natural solution to antibiotic resistance: bacteriophages “The Living Drugs”. World J Microbiol Biotechnol. 2014;30:2153–70.
Centers for Disease Control and Prevention (CDC). Antibiotic use in the United States, 2017: progress and opportunities. Atlanta: US Department of Health and Human Services; 2017.
Zucca M, Savoia D. The post-antibiotic era: promising developments in the therapy of infectious diseases. Int J Biomed Sci. 2010;6(2):77–86.
Clarke T. Drug companies snub antibiotics as pipeline threatens to run dry. Nature. 2003;425:225.
Sadhasivam S, Sinha M, Saini S, et al. Heterogeneity and antibiotic resistance in Propionibacterium acnes isolates and its therapeutic implications: blurring the lines between commensal and pathogenic phylotypes. Dermatol Ther. 2016;29(6):451–4.
Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2). https://doi.org/10.1128/microbiolspec.VMBF-0016-2015
Mendoza N, Hernandez PO, Tyring SK, Haitz KA, Motta A. Antimicrobial susceptibility of Propionibacterium acnes isolates from acne patients in Colombia. Int J Dermatol. 2013;52(6):688–92.
Luk N, Hui M, Lee H, et al. Antibiotic-resistant Propionibacterium acnes among acne patients in a regional skin centre in Hong Kong. J Eur Acad Dermatol Venereol. 2013;27(1):31–6.
Dumont-Wallon G, Moyse D, Blouin E, Dreno B. Bacterial resistance in French acne patients. Int J Dermatol. 2010;49(3):283–8.
Eady EA, Gloor M, Leyden JJ. Propionibacterium acnes resistance: a worldwide problem. Dermatology. 2003;206(1):54–6.
Dreno B, Reynaud A, Moyse D, Habert H, Richet H. Erythromycin-resistance of cutaneous bacterial flora in acne. Eur J Dermatol. 2001;11(6):549–53.
Ross JI, Snelling AM, Carnegie E, et al. Antibiotic-resistant acne: lessons from Europe. Br J Dermatol. 2003;148(3):467–78.
Tan HH, Tan AW, Barkham T, Yan XY, Zhu M. Community-based study of acne vulgaris in adolescents in Singapore. Br J Dermatol. 2007;157(3):547–51.
Kurokawa I, Nishijima S, Kawabata S. Antimicrobial susceptibility of Propionibacterium acnes isolated from acne vulgaris. Eur J Dermatol. 1999;9(1):25–8.
Nishijima S, Kurokawa I, Katoh N, Watanabe K. The bacteriology of acne vulgaris and antimicrobial susceptibility of Propionibacterium acnes and Staphylococcus epidermidis isolated from acne lesions. J Dermatol. 2000;27(5):318–23.
Burkhart C, Burkhart C. Microbiology’s principle of biofilms as a major factor in the pathogenesis of acne vulgaris. Int J Dermatol. 2003;42:925–7.
Levy R, Huang E, Roling D, Leyden J, Margolis D. Effect of antibiotics on the oropharyngeal flora in patients with acne. Arch Dermatol. 2003;139:467–71.
Mills O Jr, Thornsberry C, Cardin CW, Smiles KA, Leyden JJ. Bacterial resistance and therapeutic outcome following three months of topical acne therapy with 2% erythromycin gel versus its vehicle. Acta Derm Venereol. 2002;82(4):260–5.
Thiboutot D, Gollnick H, Bettoli V, et al. New insights into the management of acne: an update from the Global Alliance to Improve Outcomes in Acne group. J Am Acad Dermatol. 2009;60:1–50.
Grice EA. The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin Cutan Med Surg. 2014;33(2):98–103.
Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77.
Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45.
Lin L, Hong W, Ji X, Han J, Huang L, Wei Y. Isolation and characterization of an extremely long tail Thermus bacteriophage from Tengchong hot springs in China. J Basic Microbiol. 2010;50(5):452–6.
Prigent M, Leroy M, Confalonieri F, Dutertre M, DuBow M. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles. 2005;9(4):289–96.
Wittebole X, De Roock S, Opal S. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence. 2014;5(1):226–35.
Bruggemann H, Lood R. Bacteriophages infecting Propionibacterium acnes. Biomed Res Int. 2013;2013:705741.
Abedon S. Phages. In: Hyman P, Abedon S, eds. Bacteriophages in health and disease. AMCM 24 advances in molecular and cellular microbiology. Wallingford:CABI; 2012. Retrieved from https://ebookcentral.proquest.com. Accessed 7 July 2018.
O’Shea YA, Boyd EF. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol Lett. 2002;214(2):153–7.
Maiques E, Ubeda C, Tormo MA, et al. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. J Bacteriol. 2007;189(15):5608–16.
Edgar R, Friedman N, Molshanski-Mor S, Qimron U. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ Microbiol. 2012;78(3):744–51.
Lu T, Collins J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci USA. 2009;106(12):4629–34.
Brzin B. Studies on the Corynebacterium acnes. Acta Pathol Microbiol Scand. 1964;60:599–608.
Zierdt C, Webster C, Rude W. Study of the anaerobic corynebacteria. Int J Syst Evol Microbiol. 1968;18(1):33–47.
Webster G, Cummins C. Use of bacteriophage typing to distinguish Propionibacterium acne types I and II. J Clin Microbiol. 1978;7(1):84–90.
Voss JG. Differentiation of two groups of Corynebacterium acnes. J Bacteriol. 1970;101(2):392–7.
Whiteside JA, Voss JG. Incidence and lipolytic activity of Propionibacterium acnes (Corynebacterium acnes group I) and P. granulosum (C. acnes group II) in acne and in normal skin. J Investig Dermatol. 1973;60(2):94–7.
Marples RR, Leyden JJ, Stewart RN, Mills OH Jr, Kligman AM. The skin microflora in acne vulgaris. J Investig Dermatol. 1974;62(1):37–41.
Marples RR, McGinley KJ, Mills OH. Microbiology of comedones in acne vulgaris. J Investig Dermatol. 1973;60(2):80–3.
Puhvel SM, Amirian DA. Bacterial flora of comedones. Br J Dermatol. 1979;101(5):543–8.
Willner D, Furlan M, Schmieder R, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4547–53.
Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23(1):111–20.
Farrar MD, Howson KM, Bojar RA, et al. Genome sequence and analysis of a Propionibacterium acnes bacteriophage. J Bacteriol. 2007;189(11):4161–7.
Lood R, Collin M. Characterization and genome sequencing of two Propionibacterium acnes phages displaying pseudolysogeny. BMC Genom. 2011;12:198.
Brown TL, Petrovski S, Dyson ZA, Seviour R, Tucci J. The formulation of bacteriophage in a semi solid preparation for control of Propionibacterium acnes growth. PLoS One. 2016;11(3):e0151184.
Lood R, Morgelin M, Holmberg A, Rasmussen M, Collin M. Inducible siphoviruses in superficial and deep tissue isolates of Propionibacterium acnes. BMC Microbiol. 2008;8:139.
Zierdt CH. Properties of Corynebacterium acnes bacteriophage and description of an interference phenomenon. J Virol. 1974;14(5):1268–73.
Bruttin A, Brussow H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob Agents Chemother. 2005;49(7):2874–8.
Kutter E, De Vos D, Gvasalia G, et al. Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol. 2010;11(1):69–86.
Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A. Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care. 2009;18(6):237–8, 240–3.
Wright A, Hawkins C, Anggård E, Harper D. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol. 2009;34(4):349–57.
Neely K, Albright B, Zurowski M, Davis M. Development of bacteriophage therapy for the skin disease acne. In: The 108th General Meeting of the American Society for Microbiology. Boston; 2008.
O’Flaherty S, Ross R, Meaney W, Fitzgerald G, Elbreki M, Coffey A. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci fromhospitals. Appl Environ Microbiol. 2005;71:1836–42.
Schmelcher M, Donovan D, Loessner M. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7(10):1147–71.
Nelson D, Schmelcher M, Rodriguez-Rubio L, et al. Endolysins as antimicrobials. Adv Virus Res. 2012;83:299.
Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1(2):111–4.
Matsuzaki S, Yasuda M, Nishikawa H, et al. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis. 2003;187(4):613–24.
Biswas B, Adhya S, Washart P, et al. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun. 2002;70(1):204–10.
Vinodkumar CS, Neelagund YF, Kalsurmath S. Bacteriophage in the treatment of experimental septicemic mice from a clinical isolate of multidrug resistant Klebsiella pneumoniae. J Commun Dis. 2005;37(1):18–29.
Wang J, Hu B, Xu M, et al. Use of bacteriophage in the treatment of experimental animal bacteremia from imipenem-resistant Pseudomonas aeruginosa. Int J Mol Med. 2006;17:309–17.
Wang J, Hu B, Xu M, et al. Therapeutic effectiveness of bacteriophages in the rescue of mice with extended spectrum beta-lactamase-producing Escherichia coli bacteremia. Int J Mol Med. 2006;17(2):347–55.
Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brussow H. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob Agents Chemother. 2004;48(7):2558–69.
Merabishvili M, De Vos D, Verbeken G, et al. Selection and characterization of a candidate therapeutic bacteriophage that lyses the Escherichia coli O104:H4 strain from the 2011 outbreak in Germany. PLoS One. 2012;7:e52709.
Pouillot F, Chomton M, Blois H, et al. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b:H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob Agents Chemother. 2012;56(7):3568–75.
Chhibber S, Kaur T, Sandeep K. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS One. 2013;8(2):e56022.
Golkar Z, Bagasra O, Pace DG. Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J Infect Dev Ctries. 2014;8(2):129–36.
Sarker SA, McCallin S, Barretto C, et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology. 2012;434(2):222–32.
Gorski A, Miedzybrodzki R, Borysowski J, et al. Bacteriophage therapy for the treatment of infections. Curr Opin Investig Drugs. 2009;10(8):766–74.
Azeredo J, Sutherland I. The use of phages for the removal of infectious biofilm. Curr Pharm Biotechnol. 2008;9:261–6.
Alemayehu D, Casey P, McAuliffe O, et al. Bacteriophages ϕMR299-2 and ϕNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. mBio. 2012;3(2):e00029.
Alves DR, Perez-Esteban P, Kot W, et al. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb Biotechnol. 2016;9(1):61–74.
Curtin J, Donlanm R. Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob Agents Chemother. 2006;50(4):1268–75.
Fong SA, Drilling A, Morales S, et al. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front Cell Infect Microbiol. 2017;7:418.
Lehman SM, Donlan RM. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model. Antimicrob Agents Chemother. 2015;59(2):1127–37.
Comeau AM, Tétart F, Trojet S, Prère M, Krisch H. Phage–antibiotic synergy (PAS): blactam and quinolone antibiotics stimulate virulent phage growth. PLoS One. 2007;2:e799.
Coulter LB, McLean RJ, Rohde RE, Aron GM. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses. 2014;6(10):3778–86.
Hagens S, Habel A, Blasi U. Augmentation of the antimicrobial efficacy of antibiotics by filamentous phage. Microbial Drug Resist (Larchmt, NY). 2006;12(3):164–8.
Kamal F, Dennis JJ. Burkholderia cepacia complex phage-antibiotic synergy (PAS): antibiotics stimulate lytic phage activity. Appl Environ Microbiol. 2015;81(3):1132–8.
Kirby AE. Synergistic action of gentamicin and bacteriophage in a continuous culture population of Staphylococcus aureus. PLoS One. 2012;7(11):e51017.
Knezevic P, Curcin S, Aleksic V, Petrusic M, Vlaski L. Phage-antibiotic synergism: a possible approach to combatting Pseudomonas aeruginosa. Res Microbiol. 2013;164(1):55–60.
Torres-Barcelo C, Arias-Sanchez FI, Vasse M, Ramsayer J, Kaltz O, Hochberg ME. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PLoS One. 2014;9(9):e106628.
Torres-Barceló C, Hochberg M. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016;24(4):249–56.
Verma V, Harjai K, Chhibber S. Restricting ciprofloxacin-induced resistant variant formation in biofilm of Klebsiella pneumoniae B5055 by complementary bacteriophage treatment. J Antimicrob Chemother. 2009;64(6):1212–8.
Zhang QG, Buckling A. Phages limit the evolution of bacterial antibiotic resistance in experimental microcosms. Evol Appl. 2012;5(6):575–82.
Miedzybrodzki R, Fortuna W, Weber-Dabrowska B, Górski A. Phage therapy of staphylococcal infections (including MRSA) may be less expensive than antibiotic treatment. Postepy Hig Med Dosw. 2007;3:461–5.
Denou E, Bruttin A, Barretto C, Ngom-Bru C, Brussow H, Zuber S. T4 phages against Escherichia coli diarrhea: potential and problems. Virology. 2009;388(1):21–30.
Miedzybrodzki R, Borysowski J, Weber-Dabrowska B, et al. Clinical aspects of phage therapy. Adv Virus Res. 2012;83:73–121.
Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8(3):162–73.
Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B, Delattre AS, Lavigne R. Learning from bacteriophages—advantages and limitations of phage and phage-encoded protein applications. Curr Protein Pept Sci. 2012;13(8):699–722.
Acknowledgements
Funding
No funding sources were used for the development of this review.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
David E. Castillo, Sonali Nanda, and Jonette E. Keri declare have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced Digital Features
To view enhanced digital features for this article go to https://doi.org/10.6084/m9.figshare.7376021.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Castillo, D.E., Nanda, S. & Keri, J.E. Propionibacterium (Cutibacterium) acnes Bacteriophage Therapy in Acne: Current Evidence and Future Perspectives. Dermatol Ther (Heidelb) 9, 19–31 (2019). https://doi.org/10.1007/s13555-018-0275-9
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13555-018-0275-9