Abstract
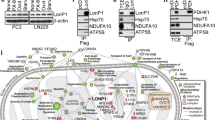
P17 is a novel neuronal protein expressed under physiological conditions only at very low levels in other tissues. Accumulating data indicate its crucial involvement in tumorigenic effects. Using molecular, cellular, and biocomputational methods, the current study unraveled p17 mode of action. Data indicate that mitochondria-associated p17 interacts with the proteins TMEM115, YPEL3, ERP44, CDK5RAP, and NNAT. Moreover, p17 drives the cell cycle into the G0/G1 phase and enhances survival of proliferating cells. Interference with p17 activities thus might become a novel option to influence also the tumor suppressor protein p53 signaling pathways for the treatment of tumors.




Similar content being viewed by others
References
Nehar S, Mishra M, Heese K. Identification and characterisation of the novel amyloid-beta peptide-induced protein p17. FEBS Lett. 2009;583(19):3247–53. doi:10.1016/j.febslet.2009.09.018.
Guo C, Zhang X, Fink SP, Platzer P, Wilson K, Willson JK, et al. Ugene, a newly identified protein that is commonly overexpressed in cancer and binds uracil DNA glycosylase. Cancer Res. 2008;68(15):6118–26. doi:10.1158/0008-5472.CAN-08-1259.
Wang LT, Lin CS, Chai CY, Liu KY, Chen JY, Hsu SH. Functional interaction of Ugene and EBV infection mediates tumorigenic effects. Oncogene. 2011;30(26):2921–32. doi:10.1038/onc.2011.16.
Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J Biol Chem. 1991;266(24):15771–81.
Heese K, Yamada T, Akatsu H, Yamamoto T, Kosaka K, Nagai Y, et al. Characterizing the new transcription regulator protein p60TRP. J Cell Biochem. 2004;91(5):1030–42. doi:10.1002/jcb.20010.
Mishra M, Akatsu H, Heese K. The novel protein MANI modulates neurogenesis and neurite-cone growth. J Cell Mol Med. 2011;15(8):1713–25. doi:10.1111/j.1582-4934.2010.01134.x.
Mishra M, Inoue N, Heese K. Characterizing the novel protein p33MONOX. Mol Cell Biochem. 2011;350(1–2):127–34. doi:10.1007/s11010-010-0690-4.
Mishra M, Heese K. P60TRP interferes with the GPCR/secretase pathway to mediate neuronal survival and synaptogenesis. J Cell Mol Med. 2011;15(11):2462–77. doi:10.1111/j.1582-4934.2010.01248.x.
Islam O, Gong X, Rose-John S, Heese K. Interleukin-6 and neural stem cells: more than gliogenesis. Mol Biol Cell. 2009;20(1):188–99. doi:10.1091/mbc.E08-05-0463.
Yokota T, Mishra M, Akatsu H, Tani Y, Miyauchi T, Yamamoto T, et al. Brain site-specific gene expression analysis in Alzheimer's disease patients. Eur J Clin Investig. 2006;36(11):820–30. doi:10.1111/j.1365-2362.2006.01722.x.
Heese K, Nagai Y, Sawada T. Induction of rat L-phosphoserine phosphatase by amyloid-beta (1-42) is inhibited by interleukin-11. Neurosci Lett. 2000;288(1):37–40.
Kelley KD, Miller KR, Todd A, Kelley AR, Tuttle R, Berberich SJ. YPEL3, a p53-regulated gene that induces cellular senescence. Cancer Res. 2010;70(9):3566–75. doi:10.1158/0008-5472.CAN-09-3219.
Berberich SJ, Todd A, Tuttle R. Why YPEL3 represents a novel tumor suppressor. Front Biosci. 2011;16:1746–51.
Tuttle R, Simon M, Hitch DC, Maiorano JN, Hellan M, Ouellette J, et al. Senescence-associated gene YPEL3 is downregulated in human colon tumors. Ann Surg Oncol. 2011;18(6):1791–6. doi:10.1245/s10434-011-1558-x.
Anelli T, Alessio M, Bachi A, Bergamelli L, Bertoli G, Camerini S, et al. Thiol-mediated protein retention in the endoplasmic reticulum: the role of ERp44. EMBO J. 2003;22(19):5015–22. doi:10.1093/emboj/cdg491.
Simmen T, Lynes EM, Gesson K, Thomas G. Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM). Biochim Biophys Acta. 2010;1798(8):1465–73. doi:10.1016/j.bbamem.2010.04.009.
Mak GW, Chan MM, Leong VY, Lee JM, Yau TO, Ng IO, et al. Overexpression of a novel activator of PAK4, the CDK5 kinase-associated protein CDK5RAP3, promotes hepatocellular carcinoma metastasis. Cancer Res. 2011;71(8):2949–58. doi:10.1158/0008-5472.CAN-10-4046.
Oyang EL, Davidson BC, Lee W, Poon MM. Functional characterization of the dendritically localized mRNA neuronatin in hippocampal neurons. PLoS One. 2011;6(9):e24879. doi:10.1371/journal.pone.0024879.
Akbari M, Otterlei M, Pena-Diaz J, Krokan HE. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 2007;145(4):1201–12. doi:10.1016/j.neuroscience.2006.10.010.
Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, et al. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–86.
Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem. 2002;277(42):39926–36. doi:10.1074/jbc.M207107200.
Aravind L, Koonin EV. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1(4):RESEARCH0007. doi:10.1186/gb-2000-1-4-research0007.
Mitra S. DNA glycosylases: specificity and mechanisms. Prog Nucleic Acid Res Mol Biol. 2001;68:189–92.
Smith ML, Seo YR. p53 regulation of DNA excision repair pathways. Mutagenesis. 2002;17(2):149–56.
Hoeijmakers JH. Genome maintenance mechanisms are critical for preventing cancer as well as other aging-associated diseases. Mech Ageing Dev. 2007;128(7–8):460–2. doi:10.1016/j.mad.2007.05.002.
Hang B, Singer B. Protein-protein interactions involving DNA glycosylases. Chem Res Toxicol. 2003;16(10):1181–95. doi:10.1021/tx030020p.
Offer H, Milyavsky M, Erez N, Matas D, Zurer I, Harris CC, et al. Structural and functional involvement of p53 in BER in vitro and in vivo. Oncogene. 2001;20(5):581–9. doi:10.1038/sj.onc.1204120.
Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20(4):914–23. doi:10.1093/emboj/20.4.914.
Inoue K, Kurabayashi A, Shuin T, Ohtsuki Y, Furihata M. Overexpression of p53 protein in human tumors. Med Mol Morphol. 2012;45(3):115–23. doi:10.1007/s00795-012-0575-6.
Lehmann BD, Pietenpol JA. Targeting mutant p53 in human tumors. J Clin Oncol. 2012;30(29):3648–50. doi:10.1200/JCO.2012.44.0412.
Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19(12):1992–2002. doi:10.1038/cdd.2012.89.
Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol. 2012;2012:170325. doi:10.1155/2012/170325.
da Costa PE, Cavalli LR, Rainho CA. Evidence of epigenetic regulation of the tumor suppressor gene cluster flanking RASSF1 in breast cancer cell lines. Epigenetics. 2011;6(12):1413–24. doi:10.4161/epi.6.12.18271.
Tuttle R, Miller KR, Maiorano JN, Termuhlen PM, Gao Y, Berberich SJ. Novel senescence associated gene, YPEL3, is repressed by estrogen in ER+ mammary tumor cells and required for tamoxifen-induced cellular senescence. Int J Cancer. 2012;130(10):2291–9. doi:10.1002/ijc.26239.
Berberich SJ. RNAi knockdown of HdmX or Hdm2 leads to new insights into p53 signaling. Cell Cycle. 2010;9(18):3640–1.
Hubertus J, Lacher M, Rottenkolber M, Muller-Hocker J, Berger M, Stehr M, et al. Altered expression of imprinted genes in Wilms tumors. Oncol Rep. 2011;25(3):817–23. doi:10.3892/or.2010.1113.
Kaushal M, Mishra AK, Sharma J, Zomawia E, Kataki A, Kapur S, et al. Genomic alterations in breast cancer patients in betel quid and non betel quid chewers. PLoS One. 2012;7(8):e43789. doi:10.1371/journal.pone.0043789.
Kuerbitz SJ, Pahys J, Wilson A, Compitello N, Gray TA. Hypermethylation of the imprinted NNAT locus occurs frequently in pediatric acute leukemia. Carcinogenesis. 2002;23(4):559–64.
Schulz R, McCole RB, Woodfine K, Wood AJ, Chahal M, Monk D, et al. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum Mol Genet. 2009;18(1):118–27. doi:10.1093/hmg/ddn322.
Evans HK, Wylie AA, Murphy SK, Jirtle RL. The neuronatin gene resides in a “micro-imprinted” domain on human chromosome 20q11.2. Genomics. 2001;77(1–2):99–104.
Uchihara T, Okubo C, Tanaka R, Minami Y, Inadome Y, Iijima T, et al. Neuronatin expression and its clinicopathological significance in pulmonary non-small cell carcinoma. J Thorac Oncol. 2007;2(9):796–801. doi:10.1097/JTO.0b013e318145af5e.
Siu IM, Bai R, Gallia GL, Edwards JB, Tyler BM, Eberhart CG, et al. Coexpression of neuronatin splice forms promotes medulloblastoma growth. Neuro Oncol. 2008;10(5):716–24. doi:10.1215/15228517-2008-038.
Xu DS, Yang C, Proescholdt M, Brundl E, Brawanski A, Fang X, et al. Neuronatin in a subset of glioblastoma multiforme tumor progenitor cells is associated with increased cell proliferation and shorter patient survival. PLoS One. 2012;7(5):e37811. doi:10.1371/journal.pone.0037811.
Contreras-Vallejos E, Utreras E, Gonzalez-Billault C. Going out of the brain: non-nervous system physiological and pathological functions of Cdk5. Cell Signal. 2012;24(1):44–52. doi:10.1016/j.cellsig.2011.08.022.
Rosales JL, Lee KY. Extraneuronal roles of cyclin-dependent kinase 5. Bioessays. 2006;28(10):1023–34. doi:10.1002/bies.20473.
Arif A. Extraneuronal activities and regulatory mechanisms of the atypical cyclin-dependent kinase Cdk5. Biochem Pharmacol. 2012;84(8):985–93. doi:10.1016/j.bcp.2012.06.027.
Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. Int J Biol Markers. 2007;22(2):108–13.
Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol Cell. 2004;15(4):621–34. doi:10.1016/j.molcel.2004.08.007.
Lu X, Nguyen TA, Appella E, Donehower LA. Homeostatic regulation of base excision repair by a p53-induced phosphatase: linking stress response pathways with DNA repair proteins. Cell Cycle. 2004;3(11):1363–6.
Macurek L, Benada J, Mullers E, Halim VA, Krejcikova K, Burdova K et al. Downregulation of Wip1 phosphatase modulates the cellular threshold of DNA damage signaling in mitosis. Cell Cycle. 2013;12(2): 251–62. doi: 10.4161/cc.23057.
Park HK, Panneerselvam J, Dudimah FD, Dong G, Sebastian S, Zhang J, et al. Wip1 contributes to cell homeostasis maintained by the steady-state level of Wtp53. Cell Cycle. 2011;10(15):2574–82.
Lowe J, Cha H, Lee MO, Mazur SJ, Appella E, Fornace Jr AJ. Regulation of the Wip1 phosphatase and its effects on the stress response. Front Biosci. 2012;17:1480–98.
Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18(20):5609–21. doi:10.1093/emboj/18.20.5609.
Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2012;493(7432):406–10. doi:10.1038/nature11725.
Acknowledgments
This study was supported by Hanyang University.