Abstract
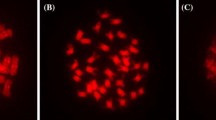
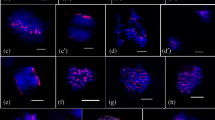
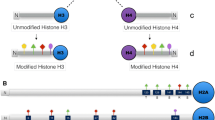
Epigenetic regulatory DNA methylation and post-translational histone modifications are considered to be highly conserved. Recent findings however suggest that chromosomal distribution of individual modifications (acetylation, methylation, phosphorylation) can differ within and between the groups of eukaryotes, indicating the obvious possibility of evolutionary divergence in reading the ‘histone code’. Orchids are known for their complex genome organization and evolution. Current trends of formation of artificial and natural polyploid orchids generally provide superior genetic variability, size, substance and form of flowers to their diploid counterparts. Here, we report the comparative chromosomal distribution of 5mC and twelve selected modified histones (methylated, acetylated and phosphorylated) between diploid and autotetraploid Dendrobium nobile at critical cytogenetic landmarks (interphase, metaphase and anaphase) of mitotic cell division to understand the underlying consequences of polyploidization on the holistic chromatin environment in higher angiosperms like orchids. Comparative in situ immuno-detection of 5-mC sites revealed increment of DNA methylation in autopolyploid D. nobile in comparison to its diploid prototype. Notably, only H3K9me1 and H3K27me2 revealed its conserved chromosomal distribution between diploid and autotetraploid D. nobile by labeling the heterochromatin and euchromatin respectively. Whereas, other epigenetic methylated and acetylated histone marks showed more or less altered/facultative distribution pattern. Histone phosphorylation did not show any alteration in chromosomal distribution between diploid and autopolyploid D. nobile. It has been concluded that polyploidization events may influence/altered the chromosomal distribution of methylated and acetylated histone marks in higher plants. This study might serve as a baseline data to understand the affect of polyploidization on epigenetic alteration associated with DNA and histone modification that ultimately play a crucial role in speciation and genome evolution of higher angiosperms like orchids.




Similar content being viewed by others
References
Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–94.
Azmi TKK, Sukma D, Aziz SA, Syukur M. Polyploidy induction of moth orchid (Phalaenopsis amabilis (L) Blume) by colchicine treatment on pollinated flowers. J Agric Sci. 2016;11:62–73.
Braszewska-Zalewska A, Bernas T, Maluszynska J. Epigenetic chromatin modifications in Brassica genomes. Genome. 2010;53:203–10.
Chen M, Ha M, Lackey E, Wang J, Chen ZJ. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics. 2008;178:1845–58.
Chen WH, Tang CY, Kao YL. Polyploidy and variety improvement of phalaenopsis orchids. Acta Hortic. 2010;878:133–8.
Chen ZJ, Ha M, Soltis D. Polyploidy: genome obesity and its consequences. New Phytol. 2007;174:717–20.
Chen ZJ, Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta. 2007;1769:295–307.
Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406.
Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–46.
De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–7.
De Paula CMP, Souza Sobrinho F, Techio VH. Chromosomal distribution of H3K4me2, H3K9me2 and 5-methylcytosine: variations associated with polyploidy and hybridization in Brachiaria (Poaceae). Plant Cell Rep. 2016;35:1359–69.
Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–94.
Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci. 2006;11:199–208.
Fuchs J, Jovtchev G, Schubert I. The chromosomal distribution of histone methylation marks in gymnosperms differs from that of angiosperms. Chromosome Res. 2008;16:891–8.
Fulnecek J, Matyásek R, Kovarík A. Faithful inheritance of cytosine methylation patterns in repeated sequences of the allotetraploid tobacco correlates with the expression of DNA methyltransferase gene families from both parental genomes. Mol Genet Genom. 2009;281:407–20.
Gerlach WL, Dyer TA. Sequence organization of the repeating units in the nucleus of wheat which contains 5S rRNA genes. Nucleic Acids Res. 1980;8:4851–65.
Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2008;7:11–7.
Griesbach RJ. Polyploidy in Phalaenopsis orchid improvement. J Hered. 1985;76:74–5.
He G, Zhu X, Elling AA, Chen L, Wang X, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33.
Hirsch CD, Wu Y, Yan H, Jiang J. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol. 2009;26:2877–85.
Houben A, Demidov D, Caperta AD, Karimi R, Agueci F, Vlasenko L. Phosphorylation of histone H3 in plants—a dynamic affair. Biochim Biophys Acta. 2007;1769:308–15.
Jackson JP, Johnson L, Jasencakova Z, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–15.
Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2010;13:153–9.
Jasencakova Z, Meister A, Schubert I. Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma. 2001;110:83–92.
Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell. 2000;12:2087–100.
Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I. Histone modifications in Arabidopsis—high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003;33:471–80.
Köhler C, Villar CB. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008;18:236–43.
Lavania UC, Srivastava S, Lavania S, Basu S, Misra NK, Mukai Y. Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J. 2012;71:539–49.
Liu B, Wendel J. Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol. 2003;29:365–79.
Masterson J. Stomatal size in fossil plants, evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–3.
Mayr C, Jasencakova Z, Meister A, Schubert I, Zink D. Comparative analysis of the functional genome architecture of animal and plant cell nuclei. Chromosome Res. 2003;11:471–84.
Mukai Y, Nakahara Y, Yamamoto M. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome. 1993;36:489–94.
Paun O, Forest F, Fay MF, Chase MW. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytol. 2009;182:507–18.
Rapp RA, Wendel JF. Epigenetics and plant evolution. New Phytol. 2005;168:81–91.
Sanei M, Pickering R, Kumke K, Nasuda S, Houben A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci USA. 2011;108:498–505.
Schatlowski N, Creasey K, Goodrich J, Schubert D. Keeping plants in shape: polycomb-group genes and histone methylation. Semin Cell Dev Biol. 2008;19:547–53.
Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–59.
Sharma SK, Dkhar J, Kumaria S, Tandon P, Rao SR. Assessment of phylogenetic inter-relationships in the genus Cymbidium (Orchidaceae) based on internal transcribed spacer region of rDNA. Gene. 2012;495:10–5.
Sharma SK, Yamamoto M, Mukai Y. Immunocytogenetic manifestation of epigenetic chromatin modification marks in plants. Planta. 2015;241:291–301.
Soltis DE, Buggs RJA, Doyle JJ, Soltis PS. What we still don’t know about polyploidy. Taxon. 2010;59:1387–403.
Soppe WJJ, Jasencakova Z, Houben A, et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–59.
Suzuki G, Shiomi M, Morihana S, Yamamoto M, Mukai Y. DNA methylation and histone modification in onion chromosomes. Genes Genet Syst. 2010;85:377–82.
Tsaftaris AS, Polidoros AN. DNA methylation and plant breeding. Plant Breed Rev. 2000;18:87–176.
Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–32.
Wang X, Elling AA, Li X, Li N, Peng Z, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–69.
Wang J, Tian L, Lee HS, Wei NE, Jiang H, et al. Genome wide non additive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–17.
Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–49.
Wood TE, Tabebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploidy speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–9.
Zhang M, Kimatu JN, Xu K, Liu B. DNA cytosine methylation in plant development. J Genet Genom. 2010;37:1–12.
Zheng B, Chen X. Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr Opin Plant Biol. 2011;14:123–9.
Acknowledgements
We thank Japan Society for the Promotion of Science (JSPS), Japan for providing postdoctoral fellowship and research grant (No. P13399 to SKS) and Grants-in-Aid for Scientific Research (C) (No. 25450006 to YM). Sincere thanks are also due to Dr. Go Suzuki and all members of Plant Molecular Genetics Laboratory, Osaka Kyoiku University, Osaka, Japan for their constant encouragement and help.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated in memory of Profs. A. K. Sharma and A. Sharma.
Rights and permissions
About this article
Cite this article
Sharma, S.K., Yamamoto, M. & Mukai, Y. Delineation of methylation and histone modification: the epigenetic regulatory marks show slightly altered distribution with the elevation in ploidy level in the orchid Dendrobium nobile. Nucleus 61, 183–193 (2018). https://doi.org/10.1007/s13237-018-0231-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-018-0231-1