Abstract
Microbial communities in hot springs at high elevations have been extensively studied worldwide. In this sense, the Indian Himalaya regions is valuable ecosystems for providing both the extreme ‘cold’ and ‘hot’ sites for exploring microbial diversity. In the present study, a total of 140 thermophilic bacteria were isolated from 12 samples collected from Manikaran and Yumthang hot springs of Indian Himalayas. The bacterial isolates were studied for phylogenetic profiling, growth properties at varying conditions and potential sources of extracellular thermostable hydrolytic enzymes such as protease, amylase, xylanase and cellulase. Based on production of extracellular hydrolases, 51 isolates from Manikaran (28) and Yumthang thermal springs (23) were selected and identified using 16S rRNA gene sequencing which included 37 distinct species of 14 different genera namely Anoxybacillus, Bacillus, Brevibacillus, Brevundimonas, Burkholderia, Geobacillus, Paenibacillus, Planococcus, Pseudomonas, Rhodanobacter, Thermoactinomyces, Thermobacillus, Thermonema and Thiobacillus. Out of 51 hydrolase producing bacteria, 24 isolates showed stability at wide range of temperature and pH treatments. In present investigation, three thermotolerant bacteria namely, Thermobacillus sp NBM6, Paenibacillus ehimensis NBM24 and Paenibacillus popilliae NBM68 were found to produced cellulase-free xylanase. These potential extracellular thermostable hydrolytic enzymes producing thermophilic bacteria have a great commercial prospect in various industrial, medical and agriculture applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbes were present in every conceivable ecological niche, includes from the tropics to the poles and hot springs to underwater hydrothermal vents. Particularly, prokaryotic communities under thermophilic habitats have been undergone the physiological adaptations to high temperature and chemical stress. Recently, these communities have attained the focus of applied research not only in terms of biotechnological prospects but also to understand the use of primitive analogues of biomolecules existed during early Earth environments (Raddadi et al. 2015; Saxena et al. 2016). Microbial communities in hot springs at low elevations have been extensively studied worldwide, however in few investigation has been done to explore the diversity in natural hot springs/thermal lakes in India. Particularly the Himalayas, though it has both extreme ‘cold’ and ‘hot’ ecosystems, but many of these valuable gene pools were remain unexplored due to inaccessible for human activities. The microbial diversity of cold and snow caped lakes such as cold deserts of Himalaya India (Sahay et al. 2013; Saxena et al. 2014, 2015; Yadav et al. 2015a, b), High-Arctic snow (Harding et al. 2011) and cold springs on the Qinghai-Tibetan Plateau, China (Yang et al. 2015) were studied. Apart from cold habitats, Himalayas have unique hot springs at high elevations, such as Soldhar and Ringigad hot springs, Uttaranchal Himalaya (Kumar et al. 2004), Chumathang hot spring, NW Indian Himalayas (Yadav 2015), Vashisht, Khirganga and Kasol hot springs (Shirkot and Verma 2015) and tattapani hot spring in NW Indian Himalayas (Priya et al. 2016).
Thermal springs are hot spots of biodiversity of microbes which can be utilized as source of novel genes, molecules and hydrolytic enzymes for agricultural, medical and industrial processes (Saxena et al. 2016). Novel thermophilic microbes have isolated and characterized from thermal extreme environments of world such as Thermotoga elfii (Ravot et al. 1995), Thermotoga hypogeal (Fardeau et al. 1997), Thermoanaerobacter uzonensis (Wagner et al. 2008) and Herbinix luporum (Koeck et al. 2016). Manikaran and Yumthang thermal springs are the hottest in the country, with a temperature range of 89–95 °C, which can be potential sources of novel genes, molecules and microorganisms. Despite intensive studies on terrestrial thermal springs, very little is known about bacterial diversity of thermal springs at high elevation. Hence, a comprehensive approach is needed to analyze the bacterial diversity of such niche, in terms of their phylogenetic profiling and its potential bioresources for extracellular hydrolytic enzymes.
Thermostable enzymes, isolated from thermophiles, have found potential commercial and industrial significance due to their inherent stability under harsh industrial processes (Verma et al. 2015) as well as stable performance at high temperature (Moraïs et al. 2016). Natural thermal environments or extreme niches are providing support to the primitive form of life; such habitats became special inherent resources of bioactive molecules for modern biotechnologists. Therefore, every country has putting research efforts towards exploration and conservation of the bio-resources available in their natural ecosystems. Potential microbes from hot springs offer a major advantage of preserving those strains for future studies and exploring them in due course for potential biotechnological applications in medical, industrial and agriculture processes. The present investigation deals with culturable thermophilic bacteria and their identification using 16S rRNA gene sequencing. Following culturable technique, isolation, characterization, phylogenetic profiling and the hydrolytic enzymes production of thermo-adapted culturable microbes was performed for samples collected from hot springs of Indian Himalayas.
Materials and methods
Study sites and sample collection
Water and sediment samples were collected from Manikaran and Yumthang hot springs, Indian Himalayas (Table 1). Manikaran hot spring is located in the Beas and Parvati valley geothermal system, at an elevation of 1760 metres with coordinate 32°02′00″N:77°20′48″E. Yumthang thermal spring is located in the North Sikkim district of Sikkim, India at an elevation of 3564 metres with coordinate 27°49′36″N:88°41′45″E. The temperatures and pH of samples were recorded at sampling sites. The temperature and pH of Manikaran hot springs ranges from 89 to 95 °C and 7.8 to 8.2 respectively whereas the temperature and pH of Yumthang hot springs ranges from 43 to 63 °C and 6.5 to 6.8 respectively. Six different samples from each hot springs collected and transferred into sterilized thermostatic bottles. All the samples were kept in sterilized thermostatic flask and were transported to the laboratory in a minimum time.
Isolation and enumeration of thermophilic bacteria
The population of culturable thermophilic bacteria in the 12 different water and sediments were enumerated through enrichment using the standard serial dilution plating technique. For enrichment, 10 mL of each water sample was inoculated directly into 50 mL of nutrient broth, where as in case of sediment samples, 5 g of sediment was mixed with 10 mL of corresponding filter sterilized water and inoculated into 50 mL of nutrient broth. All the samples were incubated at 60 °C for 2 h. Aliquots of 100 µL of each enriched samples appropriately diluted and spread on six different culture media plates namely, thermus medium (TM: Yeast extract, 4 g; polypeptone peptone, 8 g; NaCl, 2 g/L), thermus enhanced agar [TEA: Yeast extract, 2.5 g; tryptone, 2.5 g; nitrilotriacetic acid, 100 mg; CaSO4·2H2O, 40 mg; MgCl2·6H2O, 200 mg; Fe citrate (0.01 M), 0.5 mL; trace element solution, 0.5 mL (Nitrilotriacetic acid, 12.8 g; FeCl2·4H2O, 1.0 g; MnCl2·4H2O, 0.5 g; CoCl2·6H2O, 0.3 g; CuCl2·2H2O, 50 mg; Na2MoO4·2H2O, 50 mg; H3BO3, 20 mg; NiCl2·6H2O; 20 mg/L) and phosphate buffer, 0.5 mL/L (KH2PO4, 5.44 g; Na2HPO4·12H2O, 43 g/L)]; yeast extract tryptone (YET: Casein enzymic hydrolysate, 6 g; yeast extract powder, 3 g/L), Thermus thermophilus medium (TTM: Yeast extract, 4 g; proteose peptone, 8 g; NaCl, 2 g/L), nutrient agar (Peptone, 5 g; NaCl, 5 g; beef extract, 1.5 g; yeast extract, 1.5 g/L) and R2A medium (R2AM: Casein enzymic hydrolysate, 0.25 g; peptone, 0.25 g; casein acid hydrolysate, 0.5 g; yeast extract, 0.5; glucose, 0.5 g; starch soluble, 0.5 g; K2HPO4, 0.03 g; MgSO4·7H2O, 0.5 g; sodium pyruvate, 0.03 g/L) and incubated at 60 °C for 6–15 days. Colonies that appeared were purified by repeated streaking to obtain isolated colonies using respective medium plates. The pure cultures were maintained at 4 °C as slant and glycerol stock (20%) at −80 °C for further use.
Culture conditions and growth pattern of bacterial isolates
Effects of temperature and pH on growth of all the representative isolates were investigated. All the bacterial isolates were grown for 24 h at 40 °C, after that 300 µL of bacterial broth were inoculated into micro well plates having thermus liquid medium and incubated in temperature controlled Automated Microbiology Growth Analysis System (Oy Growth Curve Ab Ltd, Finland). The optical density at 600 nm was measured at regular intervals. All the isolates were analyzed in triplicates and growth curves were derived using the analysis software provided with the instrument. CFU have been recorded for each selected isolated after 24 h by inoculating 100 µL of broth on thermus medium.
Screening for thermostable enzymes producing bacterial strains
All the 140 bacterial isolates were screened for the production of extracellular thermostable hydrolases such as protease, amylase, xylanase and cellulase. The primary screening of all isolates for enzymes production was carried out as described earlier (Sahay et al. 2013). The all isolates were inoculated in the basal medium plates (Yeast extract, 1 g; KH2PO4, 1 g; MgSO4·7H2O; 0.1 g; CaCl2·H2O, 0.05 g; NaCl, 5 g; Na2CO3, 10 g and agar 15 g/L) containing starch (2.5 g/L), xylan (10 g/L), carboxy methyl cellulose (CMC) (5 g/L) and skimmed milk (10% w/v) for amylase, xylanase cellulase and protease enzymes production respectively. Purified bacterial strains were grown in 150 mL flasks containing 50 mL of nutrient broth and 5 µL of culture was spot inoculated on basal medium plates and incubated at 45, 55 and 65 °C for 3–5 days. Plates were observed for the formation of clear zone for protease. The clear zone around the colony was observed in flooded with 0.1% Congo red solution for 15 min at room temperature followed by further treatment with 1 M NaCl for cellulase and xylanase activity. The cultures were tested for amylolytic activity using basal medium supplemented with 0.025% starch. After incubation for 5 days, in each plate 0.6% KI solution was added for 30 s. The formation of zone around cultures indicated the amylase activity.
The stability of each enzyme was estimated at different temperature and pH. Selected strains were inoculated into 250 mL flasks containing 100 mL basal medium having corresponding polymer as substrate and incubated at its respective optimum growth temperature and pH for 3–5 days. An aliquot of the crude enzyme (cell free extract) was pre-incubated at different temperatures (45, 55, 65, 75 and 85 °C) for 30 min at pH 7.0 and activity was measured at its respective optimum temperature. The stability of enzyme activity (SE) at variable temperatures and pH was calculated as per the formula described in our earlier studies (Sahay et al. 2013). All the enzyme assays were conducted in triplicates and data was subjected to analysis of variance (ANOVA) using software SPSS ver. 10.
PCR amplification of 16S rRNA gene and phylogenetic analysis
Genomic DNA of selected 51 strains was extracted by the method described earlier by Sahay et al. (2011). DNA samples were subjected to PCR amplification of 16S rRNA gene using the universal primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and pH (5′-AAGGAGGTGATCCAGCCGCA-3′). The amplification was carried out in a 100 μL volume and amplification conditions were used as described earlier (Sahay et al. 2013). The PCR amplified 16S rDNA were purified with a Quiaquick purification kit (Qiagen). PCR products of partial 16S rRNA gene were sequenced with fluorescent terminators (Big Dye, Applied Biosystems) and run in 3130 × l Applied Biosystems ABI prism automated DNA sequencer at SCI Genome Chennai, India. 16S rRNA gene sequences were analysed using codon code aligner v.4.0.4. The 16S rRNA gene sequences were aligned to those of closely related bacterial species available at GenBank database using BLASTn program. Bacterial isolates were identified based on percentage of sequence similarity (≥97%) with that of a prototype strain sequence in the GenBank. The phylogenetic tree was constructed on the aligned datasets using the neighbour-joining method (NJ) implemented in the program MEGA 4.0.2 (Tamura et al. 2007). Bootstrap analysis was performed on 1000 random samples taken from the multiple alignments. The partial 16S rRNA gene sequences of 51 strains were submitted to NCBI GenBank and Accession Numbers assigned were HQ703889-90, HQ703892, HQ703893-95, HQ703897-900, HQ703902-04, HQ703909, HQ703912, HQ703918, HQ703923-24, HQ703926, HQ703930, HQ703933-34, HQ703936-37, HQ703941-42, HQ703944, HQ703950 from Manikaran hot springs and HQ851049-51, HQ851053-54, HQ851056-63, HQ851067-68, HQ851071, HQ851074, HQ851076, HQ851069, HQ851077, HQ851080-82 from Yumthang hot springs. All the 51 strains were deposited at National Bureau of Agriculturally Important Microorganisms (NBAIM) culture collection facility, Mau, Uttar Pradesh, India.
Results and discussion
The physico-chemical analysis showed specific variations among sediment and water samples collected from Manikaran and Yumthang hot springs. The samples from both springs were acidic in nature; however there was a wide variation in the temperatures of the two springs. In Manikaran, the maximum temperature was as high as 99 °C; while in Yumthang hot spring maximum temperature was 63 °C. Total six culture media were employed to retrieve the culturable bacteria from both hot springs. A total 140 bacteria were isolated from Manikaran and Yumthang hot springs. The variation was observed in bacterial species obtained from two thermal springs of distinct geographic locations. Similarly, in recent reports on natural extreme environments the culture-based approaches provided thermophiles, with highly significant for biotechnological applications (Suman et al. 2015; Huang et al. 2011; Khiyami et al. 2012; Sahoo et al. 2015; Sen and Maiti 2014; Stefanova et al. 2016; Yadav et al. 2015c).
All 140 isolates were screened for tolerance to range of temperatures and isolated were grouped into two category as thermophilic (60–90 °C) and thermotolerant (37–60 °C). Maximum isolates were grown at 45 °C; while only 19 isolates (NBM 19, NBM 48, NBM37, NBM31, NBM71, NBM75, NBM24, NBM38, NBM40, NBM49, NBY8, NBY23, NBY4, NBY33, NBY36, NBY16, NBY37, NBY28, and NBY57) showed growth at 65 °C. Further screening for their thermal tolerance at or up to 90 °C/30 min, 10 from Manikaran and 4 isolates from Yumthang hot springs showed growth (Table 2). Similarly, in case of thermophilic Bacilli obtained from hot spring showed tolerance to high temperature 70–90 °C (Lentini et al. 2007; Yadav et al. 2015c). In growth kinetic studies, the multiplication of isolates belongs to thermophilic group was observed after 12–24 h of inoculation, where as the remaining isolates from thermotolerant groups, cells growth and multiplication was observed after 20–24 h of inoculation. Our results indicated that most of the isolates from both springs showed wide pH ranges i.e. 3–8; 4–8, 4–9, 5–8 or 5–9 (Table 3). However, the isolates such as NBM24, NBM6, NBY38, NBY4, NBY5, NBY33, NBY36, NBY16, NBY26, NBY37, and NBY55, 11 isolates have showed a narrow pH range of 5–8. Out of the 51 isolates from both springs, 11 isolates from Manikaran showed tolerances to the pH range 3–8; while 23 (15 from Manikaran + 8 fromYamthang) showed growth at pH range of 4–8 however only 3 isolates from Yumthang thermal springs have showed broad pH range 4–9 and 5–9. Comparative studied has been done for each isolate for pH and temperature tolerances, it was observed that thermotolerant isolates with growth temperature range 45–65 °C showed the pH range between 3 and 9 (Table 3). Extreme environments of high temperatures can be a source for novel species of microbes, as they can tolerate extremes of temperature stress. In recent years, several studies have been conducted to look for the diversity of microbes in extreme high temperatures environments, such as geothermal areas of Deception island, in the South Shetland Archipelago (Llarch et al. 1997); Antarctic Geothermal Soils (Logan and Allan 2008); Fumarole Bay, Deception Island (Muñoz et al. 2011); permafrost volcanic sedimentary rocks (Mironov et al. 2013); Geothermal Habitats in Antarctica (Herbold et al. 2014); Hot springs of Bakreshwar, Balrampur, Chumathang and Vashist, India (Kumar et al. 2014a, b); different hot springs in India (Srivastava et al. 2014; Yadav et al. 2015c).
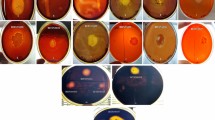
All the 140 were examined for their ability to produce extracellular hydrolytic enzymes at different temperatures and it was found that protease, amylase, cellulase and xylanase were exhibited by 30, 30, 23 and 18 strains respectively (Table 3). Among 140, 51 bacterial isolates produced one or more enzymes of protease, amylase, cellulase and xylanase (Fig. 1a, b). Among 51 bacterial strains only, 4, 6, 5, and 3 produced protease, amylase, cellulase and xylanase respectively. Among selected isolates, only 5 (9.8%) bacterial strains (NBM31, NBM71, NBY4, NBY38 and NBY44) were efficient producers of all four type thermostable enzymes (Fig. 1a, b). The selected isolates showed the stability in enzymes activities at 45 °C and/or 65 °C (Table 3). The culturable bacterial isolates of both hot springs showed thermostable enzyme activities with wide range of pH variation (Table 3). Our present results were in support with the earlier study that demonstrated thermostable cellulase production at an optimum temperature of 50 °C by Brevibacillus thermoruber (Derekova et al. 2008). There are many reports on production of thermostable cellulase and xylanase from bacterial isolates from thermal springs/high temperature habitats (Gerasimova and Kuisiene 2012; Kumar et al. 2014b; Pandey et al. 2013; Suman et al. 2015). In general the cellulases and xylanases are of special interest due to abundance of cellulose and hemicellulose in nature, moreover hydrolysis of these substrates require temperature tolerant/thermostable enzymes (Bajaj and Abbass 2011). Thermophilic microorganisms with the ability to produce proteases and amylases have also been reported from geothermal sites or thermal springs (Kumar et al. 2014b). In the present investigation, strains capable of producing thermostable proteases and amylases were identified. The thermophilic bacteria produced thermo-active enzymes, which is of great interest for both fundamental research and industrial applications. Different thermo-active enzymes production by microbes, in this study may find applications in various industries viz. food, detergents, pharmaceutical, biofuels, etc.
Xylanases are the endoactive enzymes which are generally produced in the medium containing xylan and also containing xylanase hydrolysate as the carbon source and attack the xylan chain in a random manner, causing a decrease in degree of polymerization of the substrates and liberating shorter oligomers, xylobiose and xylose. Cellulase free xylanase are of paramount significance in some of industries (Adhyaru et al. 2014; Pandey et al. 2013). Treatment with xylanase at elevated temperature disrupts the cell wall structure, facilitates lignin removal in the various stages of bleaching of paper, Therefore xylanase must lack cellulolytic activity. In present investigation, three thermotolerant bacteria namely Thermobacillus sp NBM6, Paenibacillus ehimensis NBM24 and Paenibacillus popilliae NBM68 were found to produced cellulase-free xylanase (Fig. 1a). There are many report on cellulase-free xylanase production by thermophilic bacteria e.g. Paenibacillus sp. N1 (Pathania et al. 2012) and Cellulosimicrobium cellulans CKMX1 (Walia et al. 2015). Recently, interest in xylanase has markedly increased due its wide variety of biotechnological applications such as pre-bleaching of pulp, improving the digestibility of animal feed stocks, modification of cereal-based stuffs, bioconversion of lignocellulosic material and agro-wastes to fermentable products, clarification of fruit juices and degumming of plant fibers.
16S rRNA gene sequencing and phylogenetic analysis of a selective isolates from thermal spring of Indian Himalayas, revealed that all the isolates showed >99 to 100% similarity with the sequences within the GenBank database. The phylogenetic trees were constructed to determine the affiliations for 28 and 23 bacteria isolated from thermal springs of Indian Himalayas (Fig. 2a, b). Analysis of the 16S rRNA sequences from Manikaran hot spring revealed that 25 strains belonged to Firmicutes (86%), 3 strains to Proteobacteria (10%) and 1 strain to Bacteroidetes (4%) (Fig. 2a). Yumthang hot springs bacterial isolated were grouped into four phyla namely Firmicutes (65%), Proteobacteria (30%) and Actinobacteria (4%) (Fig. 2b). Overall all identified bacteria belong to 37 distinct species of 14 different genera namely Anoxybacillus, Bacillus, Brevibacillus, Brevundimonas, Burkholderia, Geobacillus, Paenibacillus, Planococcus, Pseudomonas, Rhodanobacter, Thermoactinomyces, Thermobacillus, Thermonema and Thiobacillus.
Phylogenetic analysis of thermo-active hydrolytic enzymes producing bacteria obtained from a Manikaran hot springs; b Yumthang hot springs. The 16S rRNA gene sequence based dendrogram representing isolates were constructed by neighbor-joining method. Accession numbers of each isolate was given in parenthesis, and reference sequences having ≥97% similarity were used as taxonomic representatives. The bootstrap values (1000 replications) were indicated at the nodes
The members of phylum Firmicutes were most dominant among the identified bacteria isolated from both thermal springs. The members of phylum Bacteroidetes and proteobacteria (α, γ) were less dominant. The present investigation provides evidences that Bacillus and Bacillus derived genera were ubiquitous and predominant under harsh environments of temperatures. Similarly earlier reports also indicated the abundance of members of Firmicutes in extreme environment (Sahay et al. 2011; Verma et al. 2014, 2016a, b; Yadav et al. 2016b). Members of the Firmicutes (mainly members of families Bacillaceae and Paenibacillaceae) are widely used in agriculture as plant growth promoting and disease suppressing bacteria, besides their use in industry as a source of enzymes and in medicine (Yadav et al. 2016a, b). Further, in general the bacteria belong to the genera Bacillus and Thermus were mostly reported as aerobic, heterotrophic thermophiles and found in thermal systems with neutral to alkaline pH (Spanevello and Patel 2004). Though Thermus spp. may be predominant heterotrophs in many hot springs (Hjorleifsdottir et al. 2001) in present investigation none of the Thermus species were obtained.
In conclusion, thermo-adapted bacteria have attracted the attention of the scientific community due to their ability to produce thermostable hydrolytic extracellular enzymes. Microbial extracellular enzymes with optimal activity at high temperature provide opportunities to study the adaptation of life in thermal/high temperatures habitats and the potential for biotechnological exploitation, besides those efficient in multiple plant growth promotion traits at high temperatures. Diverse thermo-active enzymes detected in this study may be used to serve various industrial, agricultural and medicinal purposes.
References
Adhyaru DN, Bhatt NS, Modi H (2014) Enhanced production of cellulase-free, thermo-alkali-solvent-stable xylanase from Bacillus altitudinis DHN8, its characterization and application in sorghum straw saccharification. Biocatal Agric Biotechnol 3:182–190
Bajaj BK, Abbass M (2011) Studies on an alkali-thermostable xylanase from Aspergillus fumigatus MA28. 3. Biotechnology 1:161–171
Derekova A, Mandeva R, Kambourova M (2008) Phylogenetic diversity of thermophilic carbohydrate degrading bacilli from Bulgarian hot springs. World J Microbiol Biotechnol 24:1697–1702
Fardeau M-L, Ollivier B, Patel B, Magot M, Thomas P, Rimbault A, Rocchiccioli F, Garcia J-L (1997) Thermotoga hypogea sp. nov., a xylanolytic, thermophilic bacterium from an oil-producing well. Int J Syst Evol Microbiol 47:1013–1019
Gerasimova J, Kuisiene N (2012) Characterization of the novel xylanase from the thermophilic Geobacillus thermodenitrificans JK1. Microbiology 81:418–424
Harding T, Jungblut AD, Lovejoy C, Vincent WF (2011) Microbes in high arctic snow and implications for the cold biosphere. Appl Environ Microbiol 77:3234–3243
Herbold CW, McDonald IR, Cary SC (2014) Microbial ecology of geothermal habitats in Antarctica. In: Antarctic terrestrial microbiology, Springer, New York, pp 181–215
Hjorleifsdottir S, Skirnisdottir S, Hreggvidsson G, Holst O, Kristjansson J (2001) Species composition of cultivated and noncultivated bacteria from short filaments in an Icelandic hot spring at 88 °C. Microb Ecol 42:117–125
Huang Q, Dong CZ, Dong RM, Jiang H, Wang S, Wang G, Fang B, Ding X, Niu L, Li X (2011) Archaeal and bacterial diversity in hot springs on the Tibetan Plateau, China. Extremophiles 15:549–563
Khiyami MA, Serour EA, Shehata MM, Bahklia AH (2012) Thermo-aerobic bacteria from geothermal springs in Saudi Arabia. Afr J Biotechnol 11:4053–4062
Koeck DE, Hahnke S, Zverlov VV (2016) Herbinix luporum sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reactor. Int J Syst Evol Microbiol 66:4132–4137
Kumar B, Trivedi P, Mishra AK, Pandey A, Palni LMS (2004) Microbial diversity of soil from two hot springs in Uttaranchal Himalaya. Microbiol Res 159:141–146
Kumar M, Yadav AN, Tiwari R, Prasanna R, Saxena A (2014a) Evaluating the diversity of culturable thermotolerant bacteria from hour hot springs of India. J Biodivers Biopros Dev. doi:10.4172/ijbbd.1000127
Kumar M, Yadav AN, Tiwari R, Prasanna R, Saxena AK (2014b) Deciphering the diversity of culturable thermotolerant bacteria from Manikaran hot springs. Ann Microbiol 64:741–751
Lentini V, Gugliandolo C, Maugeri TL (2007) Identification of enzyme-producing thermophilic bacilli isolated from marine vents of Aeolian Islands (Italy). Ann Microbiol 57:355–361
Llarch A, Logan N, Castellví J, Prieto M, Guinea J (1997) Isolation and characterization of thermophilic Bacillus spp. from geothermal environments on Deception Island, South Shetland Archipelago. Microb Ecol 34:58–65
Logan NALA, Allan RNAN (2008) Aerobic, endospore-forming bacteria from Antarctic geothermal soils. In: Microbiology of extreme soils, Springer, New York, pp 155–175
Mironov V, Shcherbakova V, Rivkina E, Gilichinsky D (2013) Thermophilic bacteria of the genus Geobacillus from permafrost volcanic sedimentary rocks. Microbiology 82:389–392
Moraïs S, Stern J, Kahn A, Galanopoulou AP, Yoav S, Shamshoum M, Smith MA, Hatzinikolaou DG, Arnold FH, Bayer EA (2016) Enhancement of cellulosome-mediated deconstruction of cellulose by improving enzyme thermostability. Biotechnol Biofuel 9:164
Muñoz PA, Flores PA, Boehmwald FA, Blamey JM (2011) Thermophilic bacteria present in a sample from Fumarole Bay, Deception Island. Ant Sci 23:549–555
Pandey S, Singh S, Yadav AN, Nain L, Saxena AK (2013) Phylogenetic diversity and characterization of novel and efficient cellulase producing bacterial isolates from various extreme environments. Biosci Biotechnol Biochem 77:1474–1480
Pathania S, Sharma N, Verma SK (2012) Optimization of cellulase-free xylanase produced by a potential thermoalkalophilic Paenibacillus sp. N1 isolated from hot springs of Northern Himalayas in India. J Microbiol Biotechnol Food Sci 2:1–24
Priya I, Dhar M, Bajaj B, Koul S, Vakhlu J (2016) Cellulolytic activity of thermophilic bacilli isolated from Tattapani hot spring sediment in North West Himalayas. Indian J Microbiol 56:228–231
Raddadi N, Cherif A, Daffonchio D, Neifar M, Fava F (2015) Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl Microbiol Biotechnol 99:7907–7913
Ravot G, Magot M, Fardeau M-L, Patel B, Prensier G, Egan A, Garcia J-L, Ollivier B (1995) Thermotoga elfii sp. nov., a novel thermophilic bacterium from an African oil-producing well. Int J Syst Evol Microbiol 45:308–314
Sahay H, Singh S, Kaushik R, Saxena AK, Arora DK (2011) Characterization of halophilic bacteria from environmental samples from the brackish water of Pulicat Lake, India. Biologia 66:741–747
Sahay H, Babu BK, Singh S, Kaushik R, Saxena AK, Arora DK (2013) Cold-active hydrolases producing bacteria from two different sub-glacial Himalayan lakes. J Basic Microbiol 53:703–714
Sahoo RK, Subudhi E, Kumar M (2015) Investigation of bacterial diversity of hot springs of Odisha, India. Genomics Data 6:188–190
Saxena AK, Yadav AN, Kaushik R, Tyagi S, Kumar M, Prasanna R, Shukla L (2014) Use of microbes from extreme environments for the benefits of agriculture. In: Afro-Asian Congress on microbes for human and environmental health. Amity University, Noida New Delhi. doi:10.13140/RG.2.1.3479.1841
Saxena AK, Yadav AN, Kaushik R, Tyagi SP, Shukla L (2015) Biotechnological applications of microbes isolated from cold environments in agriculture and allied sectors. In: International Conference on Low temperature science and biotechnological advances. Society of low temperature biology, p 104. doi:10.13140/RG.2.1.2853.5202
Saxena AK, Yadav AN, Rajawat M, Kaushik R, Kumar R, Kumar M, Prasanna R, Shukla L (2016) Microbial diversity of extreme regions: an unseen heritage and wealth. Indian J Plant Genet Resour 29:246–248
Sen R, Maiti N (2014) Genomic and functional diversity of bacteria isolated from hot springs in Odisha, India. Geomicrobiol J 31:541–550
Shirkot P, Verma A (2015) Assessment of thermophilic bacterial diversity of thermal springs of Himachal Pradesh. ENVIS Bull Himal Ecol 23:27–34
Spanevello MD, Patel BK (2004) The phylogenetic diversity of Thermus and Meiothermus from microbial mats of an Australian subsurface aquifer runoff channel. FEMS Microbiol Ecol 50:63–73
Srivastava A, Kumar S, Kaushik R, Saxena A, Padaria J, Gupta A, Pal K, Gujar G, Sharma A, Singh P, Rai A, Kumar M, Yadav AN, Dalsania T (2014) Diversity analysis of Bacillus and other predominant genera in extreme environments and its utilization in agriculture. Technical report, 414402/C30026. doi:10.13140/2.1.1357.3927
Stefanova K, Tomova I, Tomova A, Radchenkova N, Atanassov I, Kambourova M (2016) Archaeal and bacterial diversity in two hot springs from geothermal regions in Bulgaria as demostrated by 16S rRNA and GH-57 genes. Int Microbiol 18:217–223
Suman A, Verma P, Yadav AN, Saxena AK (2015) Bioprospecting for extracellular hydrolytic enzymes from culturable thermotolerant bacteria isolated from Manikaran thermal springs. Res J Biotechnol 10:33–42
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) Software Version 4.0. Mol Biol Evol 24:1596–1599
Verma P, Yadav AN, Kazy SK, Saxena AK, Suman A (2014) Evaluating the diversity and phylogeny of plant growth promoting bacteria associated with wheat (Triticum aestivum) growing in central zone of India. Int J Curr Microbiol Appl Sci 3:432–447
Verma P, Yadav AN, Shukla L, Saxena AK, Suman A (2015) Hydrolytic enzymes production by thermotolerant Bacillus altitudinis IARI-MB-9 and Gulbenkiania mobilis IARI-MB-18 isolated from Manikaran hot springs. Int J Adv Res 3:1241–1250
Verma P, Yadav AN, Khannam KS, Kumar S, Saxena AK, Suman A (2016a) Molecular diversity and multifarious plant growth promoting attributes of Bacilli associated with wheat (Triticum aestivum L.) rhizosphere from six diverse agro-ecological zones of India. J Basic Microbiol 56:44–58
Verma P, Yadav AN, Khannam KS, Mishra S, Kumar S, Saxena AK, Suman A (2016b) Appraisal of diversity and functional attributes of thermotolerant wheat associated bacteria from the peninsular zone of India. Saudi J Biol Sci. doi:10.1016/j.sjbs.2016.01.042
Wagner ID, Zhao W, Zhang CL, Romanek CS, Rohde M, Wiegel J (2008) Thermoanaerobacter uzonensis sp. nov., an anaerobic thermophilic bacterium isolated from a hot spring within the Uzon Caldera, Kamchatka, Far East Russia. Int J Syst Evol Microbiol 58:2565–2573. doi:10.1099/ijs.0.65343-0
Walia A, Mehta P, Guleria S, Shirkot CK (2015) Modification in the properties of paper by using cellulase-free xylanase produced from alkalophilic Cellulosimicrobium cellulans CKMX1 in biobleaching of wheat straw pulp. Can J Microbiol 61:671–681
Yadav AN (2015) Bacterial diversity of cold deserts and mining of genes for low temperature tolerance. PhD thesis, BIT Mesra/IARI, New Delhi, pp 234. doi:10.1007/s11274-014-1768-z
Yadav AN, Sachan SG, Verma P, Saxena AK (2015a) Prospecting cold deserts of north western Himalayas for microbial diversity and plant growth promoting attributes. J Biosci Bioeng 119:683–693
Yadav AN, Sachan SG, Verma P, Tyagi SP, Kaushik R, Saxena AK (2015b) Culturable diversity and functional annotation of psychrotrophic bacteria from cold desert of Leh Ladakh (India). World J Microbiol Biotechnol 31:95–108. doi:10.1007/s11274-014-1768-z
Yadav AN, Verma P, Kumar M, Pal KK, Dey R, Gupta A, Padaria JC, Gujar GT, Kumar S, Suman A (2015c) Diversity and phylogenetic profiling of niche-specific Bacilli from extreme environments of India. Ann Microbiol 65:611–629
Yadav AN, Sachan SG, Verma P, Kaushik R, Saxena AK (2016a) Cold active hydrolytic enzymes production by psychrotrophic Bacilli isolated from three sub-glacial lakes of NW Indian Himalayas. J Basic Microbiol 56:294–307
Yadav AN, Sachan SG, Verma P, Saxena AK (2016b) Bioprospecting of plant growth promoting psychrotrophic Bacilli from cold desert of north western Indian Himalayas. Indian J Exp Biol 54:142–150
Yang J, Li X, Huang L, Jiang H (2015) Actinobacterial diversity in the sediments of five cold springs on the qinghai-tibet plateau. Front Microbiol 6:1345
Acknowledgements
This work was supported by Indian Council of Agricultural Research (ICAR) Network Project on Application of Microorganisms in Agriculture and Allied Sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
It is declared that there is no conflict of interest in publication of this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sahay, H., Yadav, A.N., Singh, A.K. et al. Hot springs of Indian Himalayas: potential sources of microbial diversity and thermostable hydrolytic enzymes. 3 Biotech 7, 118 (2017). https://doi.org/10.1007/s13205-017-0762-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-0762-1