Abstract
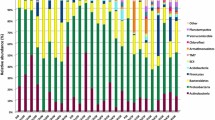
Waste disposal from paper and pulp industry is of great environmental concern. Sludge from recycled paper mills favors colonization with different microbial species due to its chemical composition. In this environment, the microbiota can express enzymes that hydrolyze celluloses and hemicelluloses, such as cellulases, β-glucosidases, and xylanases. Understanding microbiota living in such disposal may give directions to what kind of enzymes or bioproducts may be possible to obtain from this microorganism and how to deal with the waste during the process. We have accessed microbial community associated with two steps in the recycled paper sludge generation by high-throughput DNA sequencing: fibrous residue, and wastewater sludge. Bacteroidetes, Proteobacteria and Firmicutes phyla represented more than 96% of the total. From the eukaryote perspective, Vorticella was the most frequent in fibrous residue samples while Acanthamoeba was the most common on wastewater sludge samples. Among the isolates cellulase activity was detectable only from fungi isolates. Half of the bacterial isolates presented xylanase activity, and most of them in high level of activity. Both a metabarcoding and culturing approaches are valuable tools for enzymatic screening in industrial waste disposal.





Similar content being viewed by others
References
Likon, M., Trebše, P.: Recent advances in paper mill sludge management. In: Show, K.-Y., Guo, X. (eds.) Industrial Waste, pp.73–90. InTech, Rijeka (2012)
Madrid, L.M., Díaz, J.C.Q.: Ethanol production from paper sludge using Kluyveromyces marxianus. Dyna 78, 185–191 (2011)
Dwiarti, L., Boonchird, C., Harashima, S., Park, E.Y.: Simultaneous saccharification and fermentation of paper sludge without pretreatment using cellulase from Acremonium cellulolyticus and thermotolerant Saccharomyces cerevisiae. Biomass Bioenerg. 42, 114–122 (2012)
Lacour, P.: Pulp and Paper Markets in Europe—2003 Study. UNECE/FAO, Geneva (2005)
Bajpai, P.: Generation of Waste in Pulp and Paper Mills. In: Bajpai, P.. (ed.) Management of Pulp and Paper Mill Waste, pp. 9–17. Springer International Publishing, Switzerland (2015)
Chen, H., Han, Q., Daniel, K., Venditti, R., Jameel, H.: Conversion of industrial paper sludge to ethanol: fractionation of sludge and its impact. Appl. Biochem. Biotechnol. 174, 2096–2113 (2014)
Boshoff, S., Gottumukkala, L.D., van Rensburg, E., Görgens, J.: Paper sludge (PS) to bioethanol: evaluation of virgin and recycle mill sludge for low enzyme, high-solids fermentation. Bioresour. Technol. 203, 103–111 (2016)
Cavka, A., Alriksson, B., Rose, S.H., van Zyl, W.H., Jönsson, L.J.: Production of cellulosic ethanol and enzyme from waste fiber sludge using SSF, recycling of hydrolytic enzymes and yeast, and recombinant cellulase-producing Aspergillus niger. J. Ind. Microbiol. Biotechnol. 41, 1191–1200 (2014)
Kang, L., Wang, W., Pallapolu, V.R., Lee, Y.Y.: Enhanced ethanol production from de-ashed paper sludge by simultaneous saccharification and fermentation and simultaneous saccharification and co-fermentation. BioResources 6, 3791–3808 (2011)
Marques, S., Alves, L., Roseiro, J.C., Girio, F.M.: Conversion of recycled paper sludge to ethanol by SHF and SSF using Pichia stipitis. Biomass Bioenerg. 32, 400–406 (2008)
Schroeder, B.G., Zanoni, P.R.S., Magalhães, W.L.E., Hansel, F.A., Tavares, L.B.B.: Evaluation of biotechnological processes to obtain ethanol from recycled paper sludge. J. Mater. Cycles. Waste Manag. (2015). doi:10.1007/s10163-015-0445-0
Dumonceaux, T.J., Hill, J.E., Pelletier, C.P., Paice, M.G., Van Kessel, A.G., Hemmingsen, S.M.: Molecular characterization of microbial communities in Canadian pulp and paper activated sludge and quantification of a novel Thiothrix eikelboomii-like bulking filament. Can. J. Microbiol. 52, 494–500 (2006)
Riesenfeld, C.S., Schloss, P.D., Handelsman, J.: Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552 (2004)
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K-H., et al.: Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014)
Chandra, R., Singh, R.: Decolourisation and detoxification of rayon grade pulp paper mill effluent by mixed bacterial culture isolated from pulp paper mill effluent polluted site. Biochem. Eng. J. 61, 49–58 (2012)
Raj, A., Chandra, R., Reddy, M.M.K., Purohit, H.J., Kapley, A.: Biodegradation of kraft lignin by a newly isolated bacterial strain, Aneurinibacillus aneurinilyticus from the sludge of a pulp paper mill. World J. Microbiol. Biotechnol. 23, 793–799 (2007)
Zhou, J.Z., Bruns, M.A., Tiedje, J.M.: DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322 (1996)
Bates, S.T., Berg-Lyons, D., Caporaso, J.G., Walters, W.A., Knight, R., Fierer, N.: Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917 (2011)
Nolte, V., Pandey, R.V., Jost, S., Medinger, R., Ottenwalder, B., Boenigk, J., Schlotterer, C.: Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol. Ecol. 19, 2908–2915 (2010)
Schmieder, R., Edwards, R.: Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27, 863–864 (2011)
Edgar, R.C.: Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26, 2460–2461 (2010)
Edgar, R.C.: UPARSE: highly accurate OTU sequences from microbial amplicons reads. Nat. Methods. 10, 996–1000 (2013)
Cole, J.R., Wang, Q., Fish, J.A., Chai, B.L., McGarrell, D.M., et al.: Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014)
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., et al.: The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013)
Caporaso, J.G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F.D., et al.: QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336 (2010)
DeSantis, T.Z., Hugenholtz, P., Larsen, N., Roja, M., Brodie, E.L., et al.: Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006)
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., et al.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013)
Nogueira, E., Cavalcanti, M.: Cellulolytic fungi isolated from processed oats. Rev. Microbiol. 27, 7–9 (1996)
Ruijssenaars, H.J., Hartmans, S.: Plate screening methods for the detection of polysaccharase-producing microorganisms. Appl. Microbiol. Biotechnol. 55, 143–149 (2001)
Pointing, S.B.: Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Divers. 2, 17–33 (1999)
Staley, J.T., Irgens, R.L., Brenner, D.J.: Enhydrobacter aerosaccus gen. nov., sp. nov., a gas-vaculoated facultative anaerobic, heterotrophic rod. Int. J. Syst. Bacteriol. 37, 289–291 (1987)
Kawamura, T., Fujiwara, N., Naka, T., Mitani, A., Kubota, H., Tomida, J., et al.: Genus Enhydrobacter Staley et al. 1987 should be recognized as a member of the family Rhodospirillaceae within the class Alphaproteobacteria. Microbiol. Immunol. 56, 21–26 (2012)
Keely, S.P., Brinkman, N.E., Zimmerman, B.D., Wendell, D., Ekeren, K.M., et al.: Characterization of the relative importance of human- and infrastructure-associated bacteria in grey water: a case study. J. Appl. Microbiol. 119, 289–301 (2015)
Paiva, M.C., Avila, M.P., Reis, M.P., Costa, P.S., Nardi, R.M.D., Nascimento, A.M.A: The microbiota and abundance of the class 1 integron-integrase gene in tropical sewage treatment plant influent and activated sludge. PLoS One. 10, e0131532 (2015)
Premalatha, N., Gopal, N.O., Jose, P.A., Anandham, R., Kwon, S.W.: Optimization of cellulase production by Enhydrobacter sp. ACCA2 and its application in biomass saccharification. Front. Microbiol. 6, 1046 (2015)
Cao, S.-J., Deng, C.-P., Li, B.-Z., Dong, X.-Q., Yuan, H.-L.: Cloacibacterium rupense sp. nov., isolated from freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 60, 2023–2026 (2010)
Allen, T.D., Lawson, P.A., Collins, M.D., Falsen, E., Tanner, R.S.: Cloacibacterium normanense gen. nov., sp. nov., a novel bacterium in the family Flavobacteriaceae isolated from municipal wastewater. Int. J. Syst. Evol. Microbiol. 56, 1311–1316 (2006)
Ramirez, E., Robles, E., Martinez, B., Ayala, R., Sainz, G., Martinez, M.E., Gonzalez, M.E.: Distribution of free-living amoebae in a treatment system of textile industrial wastewater. Exp. Parasitol. 145, 534–538 (2014)
Kilvington, S., Price, J.: Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68, 519–525 (1990)
Santos, L.A., Fereira, V., Pereira, M.O., Nicolau, A.: Relationship between protozoan and metazoan communities and operation and performance parameters in a textile sewage activated sludge system. Eur. J. Protistol. 50, 319–328 (2014)
Cordi, L., Assalin, M.R., Ponezi, A.N., Durán, N.: Identification of microbiota for activated sludge acclimated by paper mil effluent Kraft E1 bioremediation. J Bioremed Biodeg. 3, 169 (2012)
Vaz-Moreira, I., Egas, C., Nunes, O.C., Manaia, C.M.: Culture-dependent and culture-independent diversity surveys target different bacteria: a case study in a freshwater sample. A. van Leeuw. 100, 245–257 (2011)
Meyer, T., Edwards, E.A.: Anaerobic digestion of pulp and paper mill wastewater and sludge. Water Res. 65, 321–349 (2014)
Sarto, C., Sansigolo, C.A.: Cinética da remoção dos extrativos da madeira de Eucalyptus grandis durante polpação Kraft. Acta Sci. Tech. 32, 227–235 (2010)
Sánchez, C.: Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol. Adv. 27, 185–194 (2009)
Hong, Y., Dashtban, M., Chen, S., Song, R., Qin, W.: Lignin in paper mill sludge is degraded by white-rot fungi in submerged fermentation. J. Microb. Biochem. Technol. 7, 177–181 (2015)
Niehaus, F., Bertoldo, C., Kahler, C., Antranikian, G.: Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 51, 711–729 (1999)
Gomes, E., Guez, M.A.U., Martin, N., Silva, R.: Enzimas termoestáveis: fontes, produção e aplicação industrial. Quim. Nova. 30, 136–145 (2007)
More, T.T., Yan, S., Tyagi, R.D., Surampalli, R.Y.: Potential use of filamentous fungi for wastewater sludge treatment. Bioresour. Technol. 101, 7691–7700 (2010)
Guo, J.S., Xu, Y.F.: Review of enzymatic sludge hydrolysis. J. Bioremed. Biodegrad. 2, 130 (2011)
Kumar, R., Singh, S., Singh, O.V.: Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 35, 377–391 (2008)
Sridevi, B., Charya, M.A.: Isolation, identification and screening of potential cellulase-free xylanase producing fungi. Afr. J. Biotechnol. 10, 4624–4630 (2011)
Ogbonna, A.I., Nwadiaro, P.O., Chuku, A., Ogbonna, C.I.C., Onwuliri, F.C.: Cellulase induction in three Aspergillus species isolated from Artemisia annua L. plantation soil using different cellulose substrates. J. Appl. Biotechnol. 3, 63–74 (2015)
Khokhar, I., Haider, M.S., Mushtaq, S., Mukhtar, I.: Isolation and screening of highly cellulolytic filamentous fungi. J. Appl. Sci. Environ. Manag. 16, 223–226 (2012)
Narendhirakannan, R.T., Rathore, S.S., Mannivannan, A.: Screening of cellulase producing microorganisms from lake area containing water hyacinth for enzymatic hydrolysis of cellulose. J. Adv. Sci. Res. 5, 23–30 (2014)
Romano, N., Giffré, A., Sede, S.M., Campos, E., Cataldi, A., Talia, P.: Characterization of cellulolytic activities of environmental bacterial consortia from an Argentinian native forest. Curr. Microbiol. 67, 138–147 (2013)
Maki, M.L., Broere, M., Leung, K.T., Qin, W.: Characterization of some efficient cellulase producing bacteria isolated from paper mill sludges and organic fertilizers. Int. J. Biochem. Mol. Biol. 2, 146–154 (2011)
Acknowledgements
The authors acknowledge Brazilian Agricultural Research Corporation (EMBRAPA) for funding the project “Energetic Forest-Sustainable Production and Conversion of Biomass into Energy” under which this study has been prepared; National Research Council for Scientific and Technological Development (CNPq) and Foundation for Research Support of Santa Catarina (FAPESC) for the fellowships (LBBT and KGHH), respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest declared.
Rights and permissions
About this article
Cite this article
Heinz, K.G.H., Zanoni, P.R.S., Oliveira, R.R. et al. Recycled Paper Sludge Microbial Community as a Potential Source of Cellulase and Xylanase Enzymes. Waste Biomass Valor 8, 1907–1917 (2017). https://doi.org/10.1007/s12649-016-9792-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-016-9792-x