Abstract
Inflammatory bowel disease (IBD), including ulcerative colitis and Crohn’s disease, is a chronic and relapsing inflammatory disorder of the intestine. Although its incidence is increasing globally, the precise etiology remains unclear and a cure for IBD has yet to be discovered. The most accepted hypothesis of IBD pathogenesis is that complex interactions between genetics, environmental factors, and the host immune system lead to aberrant immune responses and chronic intestinal inflammation. The human gut harbors a complex and abundant aggregation of microbes, collectively referred to as the gut microbiota. The gut microbiota has physiological functions associated with nutrition, the immune system, and defense of the host. Recent advances in next-generation sequencing technology have identified alteration of the composition and function of the gut microbiota, which is referred to as dysbiosis, in IBD. Clinical and experimental data suggest dysbiosis may play a pivotal role in the pathogenesis of IBD. This review is focused on the physiological function of the gut microbiota and the association between the gut microbiota and pathogenesis in IBD. In addition, we review the therapeutic options for manipulating the altered gut microbiota, such as probiotics and fecal microbiota transplantation.
Similar content being viewed by others
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC), which are known as inflammatory bowel diseases (IBD), are chronic and relapsing inflammatory disorders of the gastrointestinal tract [1, 2]. Over 1 million residents in the USA and 2.5 million in Europe are estimated to be suffering from IBD [3]. Although IBD is a global disease with the highest prevalence in Western countries, newly industrialized countries in Asia, the Middle East, Africa, and South America have witnessed a rapid increase in its incidence [4]. The precise etiology and pathogenesis remain to be elucidated. Genome-wide association studies have identified >200 IBD associated-susceptible genes, some of which are known to be involved or implicated in mediating host responses to gut microbiota [5]. This has evoked the possibility that gut microbiota is implicated in the pathogenesis of IBD [6, 7].
The human gut harbors 100 trillion different microbial organisms, including bacteria, viruses, fungi, and protozoa, which constitute the microbiota (also referred to as the microbial flora) [8]. Based on culture-independent molecular methods, >1000 species of bacteria reside in the gastrointestinal tract and the collective genome of intestinal microbes is estimated to contain approximately 100 times more genes than the human genome [9]. More than 99% of intestinal bacteria belong to four phyla—Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria [10, 11]. Two phyla, Bacteroidetes and Firmicutes, dominate the intestinal microbiota in healthy adults [7, 12]. The number and composition of bacteria have been reported to vary in different parts of the gastrointestinal tract. A low number and few species inhabit the stomach and the upper part of the small intestine, and the number of bacteria gradually increases from the jejunum to the colon [13].
The gut microbiota in healthy individuals is known to provide a number of health benefits to the host, relating to pathogen protection, nutrition, metabolism, and the immune system. The gut microbiota coevolves with humans, and a variety of symbiotic interactions between the human host and the microbiota is necessary to maintain human health. An unfavorable alteration of the composition and function of the gut microbiota is known as dysbiosis, which alters host−microbiota interaction and the host immune system. There is growing evidence that dysbiosis of the gut microbiota is associated with human diseases, such as IBD, irritable bowel syndrome, allergy, asthma, metabolic syndrome, and cardiovascular disease. As for IBD, many studies have reported that the composition of microbiota in IBD is altered compared with that in healthy subjects [14,15,16,17,18]. However, a direct causal relationship between dysbiosis and IBD has not been established in humans. As mentioned above, genetic predisposition is involved in the onset of IBD. However, the sharp increase in the affected population suggests that diet, which is an environmental factor, and the intestinal microbiota influenced by it are involved in the pathogenesis of IBD.
In this review, we summarize the current literature regarding the role of the gut microbiota, the association between the gut microbiota and the pathogenesis of IBD, and the potential therapeutic options for targeting the gut microbiota.
Physiological role of the gut microbiota
The benefits of the gut microbiota to the host’s physiology can be grouped into three categories—nutrition, immune development, and host defense (Fig. 1) [19]. We review these three major functions of the gut microbiota.
Nutrition
The gut microbiota supplies energy and nutrients to the host [19]. Human commensal bacteria, such as Bifidobacterium, can synthesize and supply vitamins such as vitamin K and the water-soluble B vitamins [20]. Intestinal bacteria also provide short-chain fatty acids (SCFAs; C2–C6) by fermenting resistant starch or indigestible carbohydrates (dietary fiber). The phyla Firmicutes and Bacteroidetes produce SCFAs from indigestible carbohydrates through collaboration with species specialized in oligosaccharide fermentation (e.g., Bifidobacteria) [21]. SCFAs are major anions in the colon, mainly as acetate, propionate, and butyrate [22]. Butyrate is a primary energy source for colonic epithelial cells. Butyrate is consumed mainly by the colonic epithelium, and acetate and propionate become available systemically [23]. The levels of SCFAs are significantly decreased in IBD which may be a key factor that compromises intestinal and immune homeostasis [24].
Development of the immune system
The gut microbiota plays a fundamental role in the development of the host’s immune system [25, 26]. The host immune system, in turn, shapes the structure and function of the gut microbiota [27]. Germ-free mice (deficient in the gut microbiota) exhibit impaired immune development, which is characterized by immature lymphoid tissues [28], decreased numbers of intestinal lymphocytes, and diminished levels of antimicrobial peptides [29,30,31,32] and immunoglobulin (Ig) A [33, 34]. However, reconstitution of germ-free mice with intestinal microbiota is sufficient to restore these deficits and abnormalities of the immune system [35]. One of these specific bacteria is Candidatus Arthromitis, known as segmented filamentous bacteria (SFB). The colonization of SFB alone promotes the maturation of the mucosal immune system [36,37,38]. The maturation of the host immune system is dependent on host-specific microbiota, as the immune system becomes underdeveloped in germ-free mice colonized with human microbiota [39].
The gut microbiota also modulates T-cell repertoires and regulates the T helper (Th) cell profile [40, 41]. Regulatory T cells (also called Tregs) are CD4+ T cells which have a role in regulating or suppressing other cells in the immune system [42, 43]. It has been previously demonstrated that SCFA-producing bacteria strains in Clostridium clusters IV, XIVa, and XVIII from a healthy human fecal sample induced the differentiation and expansion of colonic Tregs via butyrate production [44]. This result supports some of the clinical data. The proportion of Clostridium clusters XIVa and IV in the fecal samples is lower in IBD patients than in healthy individuals [45]. The decreased number of Faecalibacterium (F.) prausnitzii, which belong to Clostridium cluster IV, is associated with a high risk of the recurrence of CD after surgery [46].
The development of Th17 cells, which are a subpopulation of effector T cells characterized by their production of interleukin (IL)-17A, IL-17F, IL-21, and IL-22 [42], is also modulated by the gut microbiota. In germ-free or antibiotic-treated mice, the abundance of Th17 cells in the intestinal mucosa is significantly reduced. This result suggests that the gut microbiota plays a role in the development of Th17 cells. A previous report demonstrated that microbes with adhesive properties to intestinal epithelial cells, such as Citrobacter (C.) rodentium and Escherichia (E.) coli (EHEC) O157, promote the induction of Th17 cells [47].
Host defense
The gut microbiota contributes to host defense against pathogens. Animals in germ-free conditions are susceptible to infection by intestinal pathogens. An abnormality in the mucosal immune system may contribute to this susceptibility. Another mechanism against pathogens is the defect of physical and nutritional niches of the gastrointestinal tract by competitively colonizing commensal microbiota, which prevents the colonization of pathogens [19, 48]. This mechanism that competitively prevents the invasion of pathogens by commensal bacteria is called ‘colonization resistance’ [49]. The gut microbiota enhances colonization resistance to intestinal pathogens by both direct and indirect mechanisms of action. Some commensal bacteria directly inhibit intestinal pathogens by competing for nutrients or by inducing the production of inhibitory substances. Bacteroides (B.) thetaiotaomicron, which is an abundant colonic anaerobe, consumes carbohydrates used by C. rodentium, which contributes to the competitive exclusion of the pathogens from the intestinal lumen [50]. B. thuringiensis secretes a bacteriocin that directly targets spore-forming Bacilli and Clostridia, such as Clostridium difficile [51]. Commensal microbiota and microbial products protect against pathogens indirectly by activating immune responses. For instance, lipopolysaccharides and flagellin derived from the gut microbiota enhance the expression of antimicrobial peptide and RegIIIγ, from epithelial cells by stimulating Toll-like receptor 4+ stromal cells and TLR5+ CD103+ dendritic cells [52, 53]. SFB promote the secretion of IgA from B cells, the production of antimicrobial peptides and the development of Th17 cells in the intestinal mucosa [37, 54].
The gut microbiota in IBD
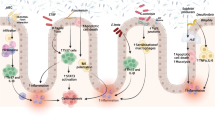
The decrease of bacteria with anti-inflammatory capacities and the increase of bacteria with inflammatory capacities are observed in patients with IBD when compared to healthy individuals [45, 55]. The most consistent changes are a reduction in the diversity of gut microbiota and the lower abundance of Firmicutes (Fig. 2) [7, 45, 46, 56]. Increases in abundance of Proteobacteria and Bacteroidetes have been reported, but reductions have also been reported [45, 56]. F. prausnitzii, which belongs to Clostridium cluster IV, has been reported to have an anti-inflammatory effect by producing butyrate. It has been demonstrated that F. prausnitzii, Blautia faecis, Roseburia inulinivorans, Ruminococcus torques, and Clostridium lavalense are decreased in patients with CD when compared to healthy subjects [15, 18] and that the number of F. prausnitzii is correlated with the risk of relapse of ileal CD after surgery. The defect of colonization of F. prausnitzii was observed in UC patients during remission and the recovery of the F. prausnitzii population after relapse is associated with the maintenance of clinical remission [57]. In addition, Sokol et al. showed that human peripheral blood mononuclear cells stimulated with F. prausnitzii induce the production of IL-10 and inhibit the production of inflammatory cytokines, such as IL-12 and IFN-γ [58]. Furthermore, a significant decrease of Roseburia spp. was shown in the gut microbiota of healthy individuals with a high genetic risk for IBD.
Dysbiosis in IBD and pathological outcomes of dysbiosis. An unfavorable alteration of the composition and variety of the gut microbiota is termed dysbiosis. Dysbiosis is observed in IBD patients. Dysbiosis affects the host immune system and barrier integrity, resulting in chronic inflammation and aberrant immune responses
In contrast, a relative increase in Proteobacteria, mainly E. coli, was observed in CD patients, in particular, on mucosa-associated microbiota compared to fecal samples [16, 59,60,61,62,63,64,65]. CD-associated E. coli with pro-inflammatory properties is adhesion-invasive E. coli (AIEC), which was originally isolated from adult CD patients [55]. It has been reported that the number of AIEC increased in about 38% of patients with active CD compared to 6% in healthy subjects [66]. The increase of pathogenic bacteria with the ability to adhere to the intestinal epithelium affects the permeability of the intestine, alters the diversity and composition of gut microbiota, and induces inflammatory responses by regulating the expression of inflammatory genes, consequently leading to the induction of intestinal inflammation [67]. In addition, fluorescence in situ hybridization analyses have shown an increased abundance of mucosa-associated bacteria in IBD [68,69,70]. This may be caused by the increased number of mucolytic bacteria, such as Runinococcus gnavas and Ruminococcus torques in IBD patients [70].
The production of metabolites affected by the disruption of gut microbiota is associated with the pathogenesis of IBD. For instance, the concentration of SCFAs has been reported to decrease in IBD patients, as a result of butyrate-producing bacteria, such as F. prausnizzi and Clostridium clusters IV, XIVa, XVIII [18]. The decreased production of SCFAs affects the differentiation and expansion of Treg cells and the growth of epithelial cells [44], which play an important role in maintaining intestinal homeostasis. On the other hand, the number of sulfate-reducing bacteria, such as Desulfovibrio, is higher in IBD patients [71, 72], resulting in the production of hydrogen-sulfate that damages intestinal epithelial cells and induces mucosal inflammation [71, 73]. Collectively, these data strongly indicate that the alteration of gut microbiota is associated with the pathogenesis of IBD.
The effects of medication for IBD on gut microbiota
Medication for IBD also affects the composition of gut microbiota. Mesalazine reduces fecal bacteria and the concentration of mucosal adherent bacteria when compared to untreated patients [74, 75]. Mesalazine inhibits the growth of Mycobacterium avium subspecies paratuberculosis, which has been reported to be intimately linked to the etiology of CD [76], in a dose-dependent manner in vitro [77]. Another study has reported that mesalazine downregulates the gene expression that is associated with bacterial invasiveness and antibiotic resistance in Salmonella enterica serovar Typhimurium [78], which could promote the onset of IBD after its infection [79, 80]. Furthermore, mesalazine inhibits the growth of sulfate-reducing bacteria and suppresses sulfide production in IBD patients [81, 82]. Indeed, IBD patients who do not use mesalazine have higher fecal sulfide levels [81].
Anti-tumor necrosis factor (TNF)-α antibody therapy has been reported to affect the gut microbiota. For instance, the abundance of F. prausnitzii, which can exert anti-inflammatory effects via metabolites [12], increased in responders during the induction of anti-TNF-α antibody therapy [83]. Another study reported that the relative number of E. coli in CD had significantly decreased and that of F. prausnitzii had increased when they were measured 3 months after the start of anti-TNF-α antibody therapy [84].
There is little information available about the effects of thiopurine and glucocorticoids on the gut microbiota. The treatment of thiopurine increases the concentration and adherence of mucosal bacteria [85]. It has been found that thiopurine inhibits the growth of Mycobacterium avium subspecies paratuberculosis in vitro [86, 87]. Another study has reported that thiopurine use significantly reduced the bacterial diversity and richness in fecal samples in IBD when compared to other drugs, including anti-TNF-α antibodies, mesalazine, and corticosteroids [88].
The effect of glucocorticoids on the gut microbiota has been investigated in animal models but not in humans. In mouse models, glucocorticoids increase the abundance of Bifidobacterium and Lactobacillus, while eliminating the presence of Mucispirillum, known as mucin degrader, under normal and inflammatory conditions [89]. Another study using healthy dogs has reported that no significant change was observed in the gut microbiota due to the administration of glucocorticoids [90].
Collectively, medication for IBD could change or modulate the gut microbiota. However, reliable evidence is poor. Furthermore, it remains unknown whether the change in gut microbiota is the result of medication or the consequence of the improvement of intestinal inflammation.
Therapeutic strategy for targeting dysbiosis
Probiotics
Probiotics are living microorganisms that exert beneficial effects on the host by modulating the intestinal microbiota [91]. Many probiotics have been examined for their efficacy in IBD with the aim of modulating the microbiota and relieving intestinal dysbiosis. It has been demonstrated that some bacteria, such as Lactobacillus, Bifidobacterium and Streptococcus, have a clinical effect on gastrointestinal inflammation. A large clinical trial was conducted to investigate the efficacy of E. coli Nissle 1917, a nonpathogenic strain, on maintaining remission of UC. E. coli Nissle 1917 achieved comparable efficacy and safety outcomes to salicylate in the maintenance of remission in UC patients [92]. Studies using VSL#3, a probiotic mixed with 4 Lactobacilli (L.) (L. casei, L. acidophilus, L. delbrueckii subsp., Bulgaricus), 3 Bifidobacteria (B.) (B. longum, B. breve, B. infantis) and a Streptococcus (Streptococcussalivarius subsp. thermophilus), have yielded the most available evidence in IBD patients. A clinical study found that VSL#3 was effective in inducing remission in patients with mild-to-moderately active UC [93, 94]. In addition, in a small cohort study, VSL#3 was found to be effective in maintenance of remission [93]. A meta-analysis has recently shown that VSL#3 with standard treatment achieved better outcomes when compared to standard treatment alone in the induction of remission and response [95]. A randomized study to investigate the efficacy of Lactobacillus GG was conducted in quiescent UC. Lactobacillus GG was safe and more effective than standard treatment with mesalazine for maintaining remission in UC patients [96]. Several systematic reviews and meta-analyses have addressed the role of probiotics in UC without conclusive outcomes, and a Cochrane review showed that probiotics were no more effective than placebo or standard treatment in inducing the remission of UC [97].
In contrast, there is limited evidence available for the efficacy of probiotics in CD patients. A randomized, double-blind, placebo-controlled study investigated the efficacy of L. johnsonii in CD patients who underwent surgical resection of the intestine; however, L. johnsonii did not have a sufficient effect on the suppression of recurrence rate [98]. Other probiotics, such as E. coli Nissle 1917 [99] and Saccharomyces boulardi [100], did not have a significant effect on remission rate in CD patients. A Cochrane review reported that probiotics have no advantage over placebo in the maintenance and induction of CD, and suggested that well-designed randomized clinical trials (RCTs) should be conducted to address the efficacy of probiotics in the treatment of CD [101].
Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) aims to restore the intestinal microbiota in diseased individuals by transferring intestinal microbiota of healthy donors. FMT has been clinically adapted to recurrent Clostridium difficile infection (CDI), and the efficacy of FMT for CDI has been established with a high cure rate of >90% in clinical trials [102, 103]. The success of FMT in treating CDI has raised the possibility that FMT may be beneficial in other diseases associated with dysbiosis. Therefore, FMT has recently attracted attention as a new therapeutic strategy in IBD.
The number of clinical studies on the efficacy and safety of FMT for IBD has increased, and the evidence for the effect of FMT on IBD has accumulated accordingly. Initial data to support the role of FMT in treating IBD came from case reports. Recent reports of the use of FMT in adult patients with UC yielded mostly no or limited effects. Subsequently, two randomized placebo-controlled clinical trials in patients with active UC were recently published in 2015 (Table 1) [104, 105]. Moayyedi et al. [104] reported that 75 subjects received FMT or placebo (water) by retention enema for 6 weeks and that the remission rate (Mayo Score of <3 with an endoscopic subscore of 0 at week 7) of the FMT group was significantly higher than the placebo group (9/38 [24%] vs 2/37 [5%]; P = 0.03). On the other hand, in the clinical trial reported by Rossen et al., 50 patients with mild-to-moderately active UC were treated with either donor stool (FMT group) or autologous FMT (placebo group) delivered via nasoduodenal tube at day 0 and 3 weeks later, and there was no significant difference in the remission rate (clinical remission combined with a ≥1-point decrease in the Mayo endoscopic score at week 12) between the two groups [105]. Notably, the two above-mentioned clinical trials were stopped early for futility by the Data Monitoring and Safety Committee. More recently, another two RCTs in patients with mild-to-moderately active UC were reported in 2017 (Table 1). Paramsothy et al. [106] reported that 85 patients were randomly assigned FMT or placebo, and that the primary outcome was defined as steroid-free clinical remission with endoscopic remission or response (Mayo score of ≤2, all subscores ≤1, and ≥1-point reduction in endoscopy subscore) at week 8. The rate of the primary outcome of the FMT group was significantly higher than the placebo group (11/41 [27%] vs 3/40 [8%], P = 0.02). Costello et al. [107] reported that 73 patients with active UC were enrolled and randomly allocated to FMT or autologous FMT (placebo). Prepared donor stool was administered via colonoscopy on day 0 followed by two enemas by day 7. The remission rate (steroid-free remission, Mayo score ≤2 and endoscopic subscore ≤1) of FMT was significantly higher than that of the placebo group (12/38 [32%] vs 3/35 [9%]; P = 0.02). Paramsothy et al. [108] recently published a systematic review and meta-analysis of 53 studies that included 661 IBD patients who were treated by FMT. They found that the clinical remission rate of UC patients was 36%. They also found a significant benefit in clinical remission in UC for four above-mentioned RCTs (pooled odds ratios = 2.89, 95% CI = 1.36–6.13, P = 0.006).
FMT has also been applied to CD patients. All reports of FMT for CD patients published to date are case reports or cohort studies. A recent systematic review and meta-analysis of 11 studies (four case reports and seven prospective uncontrolled cohort studies) on FMT for CD that included 83 CD patients demonstrated an overall clinical remission rate of 50.5% (42/83) [108]. The authors indicated that publication bias was observed in this meta-analysis. In the future, RCTs should be conducted to evaluate the efficacy of FMT on CD patients.
It is obvious that FMT is not as effective in IBD as it is in CDI. CDI develops because of the disruption of gut microbiota using antibiotics followed by an overgrowth of Clostridium difficile. Therefore, the treatment with FMT shows high cure rates for recurrent CDI, regardless of donor, recipient, and delivery method [102]. On the other hand, various factors such as microbial, genetic, immunologic, and environmental factors are involved in the pathogenesis of IBD. The interplay between the host and gut microbiota in IBD is more complicated than that in CDI. In addition, the protocol of FMT, including the criteria of donor selection, patient pretreatment and administration route, is different in each clinical trial, which results in different effects. Finally, the most important issue is that dysbiosis is associated with the pathogenesis of IBD, but it is not clear whether this is a cause or a result of the inflammatory process. It is possible that all of these factors might contribute to the low clinical efficacy of FMT for IBD compared to that of FMT for CDI.
The factors involved in the response to FMT in UC patients remain unclear. An RCT conducted by Moayyedi et al. [104] found that early diagnosis of UC in patients might result in better outcomes with FMT. This suggests the possibility that there is a window of opportunity for FMT after diagnosis of UC. Paramsothy et al. [106] suggested that less severe endoscopic inflammation may be a potential predictor of FMT response. Moreover, the increase of Clostridium clusters IV and XVIII and the high microbial diversity in patients were identified as predictors of FMT response [106]. Conversely, the presence of Fusobacterium spp. and Sutterella spp. in patients were associated with non-response to FMT [106].
Based on the clinical data, FMT may remain in clinical trials but not in clinical practice. More high-quality clinical studies are necessary to optimize the protocol for FMT and determine the efficacy of FMT in IBD and long-term safety in the future.
Future directions
The recent application of DNA sequencing technology and the availability of genetically engineered animal models and gnotobiotic animals have resulted in great advances in understanding the interaction between the gut microbiota and the host. The gut microbiota plays an important role in the maintenance of intestinal homeostasis and the development and activation of the host immune system. The microbiota shapes the development of the immune system, and the immune system, in turn, shapes the gut microbiota. Moreover, the gut microbiota influences the susceptibility to disease, and even the metabolic function of the host. These recent advances in our understanding of the gut microbiota have revealed that the unfavorable alteration of the gut microbiota, i.e., dysbiosis, is strongly associated with the pathogenesis of IBD. They have also contributed to the development of novel therapeutic options that selectively target dysbiosis in IBD. For instance, FMT has attracted attention as a new therapeutic strategy for IBD. However, although it has significant effects in the treatment of CDI, its efficacy in IBD remains controversial. There are many factors that should be taken into consideration in order to increase the success rate of FMT in IBD, such as disease state, donor selection, and the standardizing of the fecal microbiota processing protocol. In addition, a recent study has raised a question about the role of the enteric virome in FMT for IBD. Zou et al. [109] reported that the role of Caudovirales bacteriophages was potentially associated with the efficacy of FMT for CDI. Therefore, the gut virome, as well as the gut microbiota, should be taken into consideration when evaluating the efficacy of FMT for IBD. In the future, discoveries in the field of the gut microbiota will surely contribute to the development of novel therapeutic strategies in IBD.
References
Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621.
Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–17.
Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–7.
Kaplan GG, Ng SC. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology. 2017;152(2):313–321.e2.
Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86.
Goldsmith JR, Sartor RB. The role of diet on intestinal microbiota metabolism: downstream impacts on host immune function and health, and therapeutic implications. J Gastroenterol. 2014;49(5):785–98.
Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol. 2015;50(5):495–507.
Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–95.
Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65.
Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8.
Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–51.
Andoh A. Physiological role of gut microbiota for maintaining human health. Digestion. 2016;93(3):176–81.
Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–94.
Andoh A, Sakata S, Koizumi Y, et al. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13(8):955–62.
Fujimoto T, Imaeda H, Takahashi K, et al. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J Gastroenterol Hepatol. 2013;28(4):613–9.
Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S, Inatomi O, Bamba S, Sugimoto M, Kawahara M, Naito Y, Andoh A. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 2017. https://doi.org/10.1007/s00535-017-1384-4.
Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152(2):327–339.e4.
Takahashi K, Nishida A, Fujimoto T, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion. 2016;93(1):59–65.
O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–93.
LeBlanc JG, Laino JE, del Valle MJ, et al. B-group vitamin production by lactic acid bacteria–current knowledge and potential applications. J Appl Microbiol. 2011;111(6):1297–309.
Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–9.
Sun M, Wu W, Liu Z, et al. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52(1):1–8.
Pomare EW, Branch WJ, Cummings JH. Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J Clin Invest. 1985;75(5):1448–54.
Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275–83.
Falk PG, Hooper LV, Midtvedt T, et al. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62(4):1157–70.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23.
Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology. 2014;146(6):1477–88.
Bouskra D, Brezillon C, Berard M, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456(7221):507–10.
Ayabe T, Satchell DP, Pesendorfer P, et al. Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem. 2002;277(7):5219–28.
Cash HL, Whitham CV, Behrendt CL, et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–30.
Hooper LV, Stappenbeck TS, Hong CV, et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–73.
Putsep K, Axelsson LG, Boman A, et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. 2000;275(51):40478–82.
Hapfelmeier S, Lawson MA, Slack E, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328(5986):1705–9.
Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4(6):478–85.
Umesaki Y, Okada Y, Matsumoto S, et al. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39(8):555–62.
Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–89.
Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98.
Umesaki Y, Setoyama H, Matsumoto S, et al. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67(7):3504–11.
Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–93.
Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69(5):1046s–51s.
Shanahan F. The host-microbe interface within the gut. Best Pract Res Clin Gastroenterol. 2002;16(6):915–31.
Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140(6):845–58.
Sakaguchi S, Yamaguchi T, Nomura T, et al. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–87.
Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–6.
Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104(34):13780–5.
Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7.
Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163(2):367–80.
Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904.
Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13(11):790–801.
Kamada N, Kim YG, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336(6086):1325–9.
Huang T, Zhang X, Pan J, et al. Purification and characterization of a novel cold shock protein-like bacteriocin synthesized by Bacillus thuringiensis. Sci Rep. 2016;6:35560.
Kinnebrew MA, Ubeda C, Zenewicz LA, et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201(4):534–43.
Brandl K, Plitas G, Mihu CN, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–7.
Talham GL, Jiang HQ, Bos NA, et al. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67(4):1992–2000.
Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3(6):417–27.
Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–11.
Varela E, Manichanh C, Gallart M, et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther. 2013;38(2):151–61.
Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–6.
Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1(5):403–18.
Kotlowski R, Bernstein CN, Sepehri S, et al. High prevalence of Escherichia coli belonging to the B2 + D phylogenetic group in inflammatory bowel disease. Gut. 2007;56(5):669–75.
Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127(1):80–93.
Martinez C, Antolin M, Santos J, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103(3):643–8.
Mylonaki M, Rayment NB, Rampton DS, et al. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11(5):481–7.
Neut C, Bulois P, Desreumaux P, et al. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am J Gastroenterol. 2002;97(4):939–46.
Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122(1):44–54.
Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127(2):412–21.
Ahmed I, Roy BC, Khan SA, et al. Microbiome, metabolome and inflammatory bowel disease. Microorganisms. 2016;4(2):20.
van der Waaij LA, Harmsen HJ, Madjipour M, et al. Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA-based fluorescent probes: commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm Bowel Dis. 2005;11(10):865–71.
Schultsz C, Van Den Berg FM, Ten Kate FW, et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117(5):1089–97.
Png CW, Linden SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105(11):2420–8.
Loubinoux J, Bronowicki JP, Pereira IA, et al. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol. 2002;40(2):107–12.
Zinkevich VV, Beech IB. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol Ecol. 2000;34(2):147–55.
Rowan F, Docherty NG, Murphy M, et al. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53(11):1530–6.
Andrews CN, Griffiths TA, Kaufman J, et al. Mesalazine (5-aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2011;34(3):374–83.
Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43(7):3380–9.
Naser SA, Sagramsingh SR, Naser AS, et al. Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J Gastroenterol. 2014;20(23):7403–15.
Greenstein RJ, Su L, Shahidi A, et al. On the action of 5-amino-salicylic acid and sulfapyridine on M. avium including subspecies paratuberculosis. PLoS One. 2007;2(6):e516.
Kaufman J, Griffiths TA, Surette MG, et al. Effects of mesalamine (5-aminosalicylic acid) on bacterial gene expression. Inflamm Bowel Dis. 2009;15(7):985–96.
Gradel KO, Nielsen HL, Schonheyder HC, et al. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137(2):495–501.
Schultz BM, Paduro CA, Salazar GA, et al. A potential role of salmonella infection in the onset of inflammatory bowel diseases. Front Immunol. 2017;8:191.
Pitcher MC, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut. 2000;46(1):64–72.
Edmond LM, Hopkins MJ, Magee EA, et al. The effect of 5-aminosalicylic acid-containing drugs on sulfide production by sulfate-reducing and amino acid-fermenting bacteria. Inflamm Bowel Dis. 2003;9(1):10–7.
Magnusson MK, Strid H, Sapnara M, et al. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J Crohns Colitis. 2016;10(8):943–52.
Busquets D, Mas-de-Xaxars T, Lopez-Siles M, et al. Anti-tumour necrosis factor treatment with adalimumab induces changes in the microbiota of Crohn’s Disease. J Crohns Colitis. 2015;9(10):899–906.
Swidsinski A, Loening-Baucke V, Bengmark S, et al. Azathioprine and mesalazine-induced effects on the mucosal flora in patients with IBD colitis. Inflamm Bowel Dis. 2007;13(1):51–6.
Shin SJ, Collins MT. Thiopurine drugs azathioprine and 6-mercaptopurine inhibit Mycobacterium paratuberculosis growth in vitro. Antimicrob Agents Chemother. 2008;52(2):418–26.
Greenstein RJ, Su L, Haroutunian V, et al. On the action of methotrexate and 6-mercaptopurine on M. avium subspecies paratuberculosis. PLoS One. 2007;2(1):e161.
Wills ES, Jonkers DM, Savelkoul PH, et al. Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation. PLoS One. 2014;9(3):e90981.
Huang EY, Inoue T, Leone VA, et al. Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflamm Bowel Dis. 2015;21(5):963–72.
Igarashi H, Maeda S, Ohno K, et al. Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLoS One. 2014;9(9):e107909.
Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics—approaching a definition. Am J Clin Nutr. 2001;73(2 Suppl):361s–4s.
Kruis W, Fric P, Pokrotnieks J, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53(11):1617–23.
Sood A, Midha V, Makharia GK, et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol. 2009;7(11):1202–1209.e1.
Tursi A, Brandimarte G, Giorgetti GM, et al. Low-dose balsalazide plus a high-potency probiotic preparation is more effective than balsalazide alone or mesalazine in the treatment of acute mild-to-moderate ulcerative colitis. Med Sci Monit. 2004;10(11):Pi126–31.
Mardini HE, Grigorian AY. Probiotic mix VSL#3 is effective adjunctive therapy for mild to moderately active ulcerative colitis: a meta-analysis. Inflamm Bowel Dis. 2014;20(9):1562–7.
Zocco MA, dal Verme LZ, Cremonini F, et al. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 2006;23(11):1567–74.
Mallon P, McKay D, Kirk S, et al. Probiotics for induction of remission in ulcerative colitis. Cochrane Database Syst Rev. 2007;4:Cd005573.
Marteau P, Lemann M, Seksik P, et al. Ineffectiveness of Lactobacillus johnsonii LA1 for prophylaxis of postoperative recurrence in Crohn’s disease: a randomised, double blind, placebo controlled GETAID trial. Gut. 2006;55(6):842–7.
Malchow HA. Crohn’s disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn’s disease? J Clin Gastroenterol. 1997;25(4):653–8.
Guslandi M, Mezzi G, Sorghi M, et al. Saccharomyces boulardii in maintenance treatment of Crohn’s disease. Dig Dis Sci. 2000;45(7):1462–4.
Butterworth AD, Thomas AG, Akobeng AK. Probiotics for induction of remission in Crohn’s disease. Cochrane Database Syst Rev. 2008;3:Cd006634.
Quraishi MN, Widlak M, Bhala N, et al. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther. 2017;46(5):479–93.
van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–15.
Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149(1):102–109.e6.
Rossen NG, Fuentes S, van der Spek MJ, et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology. 2015;149(1):110–118.e4.
Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389(10075):1218–28.
Costello SP, Waters O, Bryant RV, et al. Short duration, low intensity, pooled fecal microbiota transplantation induces remission in patients with mild-moderately active ulcerative colitis: a randomised controlled trial. Gastroenterology. 2017;152(5):S198–9. https://doi.org/10.1016/S0016-5085(17)30989-1.
Paramsothy S, Paramsothy R, Rubin DT, et al. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2017;11(10):1180–99.
Zuo T, Wong SH, Lam K, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2017. https://doi.org/10.1136/gutjnl-2017-313952.
Acknowledgements
This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15K08967, 15K19322), a grant for the Intractable Diseases from the Ministry of Health, Labor and Welfare of Japan (067), a Grant from the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development, AMED (15AeK0109047h0002), and a grant from the Smoking Research Foundation (1848).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors disclose no conflict of interest.
Rights and permissions
About this article
Cite this article
Nishida, A., Inoue, R., Inatomi, O. et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 11, 1–10 (2018). https://doi.org/10.1007/s12328-017-0813-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12328-017-0813-5