Abstract
Equol is a daidzein (a phytoestrogen isoflavone) metabolite of gut bacteria, and the ability to produce equol varies between individuals and reduces the risks of several diseases. We tested the effects of equol production on health in Koreans and identified the genetic factors that determine the equol-producing phenotype. In 1391 subjects, the equol-producing phenotype was determined, based on measurements of serum equol concentrations. The anthropometric and blood biochemical measurements between equol producers and nonproducers were analyzed by LC-MS/MS. Genetic factors were identified in a genomewide association study (GWAS), and the interaction between genetic factors and the equol-producing phenotype was examined. We observed that 70.1 % of the study population produced equol. Blood pressure was significantly lower in equol producers (beta ± SE = −1.35 ± 0.67, p = 0.045). In our genomewide association study, we identified 5 single-nucleotide polymorphisms (p < 1 × 10−5) in HACE1. The most significant SNP was rs6927608, and individuals with a minor allele of rs6927608 did not produce equol (odds ratio = 0.57 (95 % CI 0.45–0.72), p value = 2.5 × 10−6). Notably, the interaction between equol production and the rs6927608 HACE1 SNP was significantly associated with systolic blood pressure (p value = 1.3 × 104). Equol production is linked to blood pressure, and HACE1, identified in our (GWAS), might be a determinant of the equol-producing phenotype.
Similar content being viewed by others
Introduction
Asians regularly consume soy-containing foods, such as tofu, doenjang soup, and cheonggukjang (Kwon et al. 2010; Song et al. 2006), and have a lower rate of cardiovascular disease and certain types of cancers than Western populations (DellaPenna 1999; Sakar and Li 2002; reviewed in Andres et al. 2011). However, the mechanisms by which these benefits are effected are unknown—benefits, however, have been debated, as demonstrated by the controversial effects of isoflavone on bone density in postmenopausal women (Ma et al. 2008; Brink et al. 2008; Alekel et al. 2010).
Soy-based foods contain bioactive compounds, such as isoflavones (genistein, daidzein, and glycitein) (Setchell 1998). Daidzein can be metabolized by intestinal bacteria to equol (reviewed in Rowland et al. 1999 and Atkinson et al. 2005). Notably, daidzein metabolism differs between individuals, contributing to variations in isoflavone profiles (Heinonen et al. 1999). Daidzein-metabolizing phenotypes are divided into two groups: equol producers and nonproducers (Setchell et al. 2002). The prevalence of equol producers among Western adults is 20 to 35 % compared with 50 to 55 % among Asian adults (Yuan et al. 2007); a Korean study has reported a rate of 59 % (Akaza et al. 2004).
Recent studies indicate that daidzein-metabolizing phenotypes are associated with risk biomarkers for several diseases (Atkinson et al. 2004a, b). In a Chinese report, the prevalence of equol producers was lower in breast cancer cases than controls (Dai et al. 2002). Three prostate case–control studies in Japanese and Koreans have suggested that serum equol levels correlate with a lower risk of prostate cancer (Akaza et al. 2002, 2004; Ozasa et al. 2004). Additionally, based on its antioxidant and anti-inflammatory activities, as shown in in vitro studies, equol is linked to atherosclerosis and cardiovascular disease (Setchell and Clerici 2010 as a review). Yet, controversy remains over whether there is any advantage in producing equol as a consequence of soy isoflavone intakes.
Interindividual differences in daidzein-metabolizing phenotypes are attributed to disparities in the gut microbial environment (Atkinson et al. 2004a). Host defense systems must distinguish commensal organisms from episodic pathogens and regulate the ensuing responses precisely (O’Hara and Shanahan 2006). Thus, it is conceivable that daidzein-metabolizing phenotypes are governed tightly by the host defense system, and equol synthesis from daidzein might be regulated by intestinal equol-producing bacteria. In this study, we hypothesized that host genetic factors influence the ability to develop and maintain daidzein-metabolizing bacteria and that the resulting equol modulates the host health index.
To examine the relationship between equol production and host health, 1391 samples from a general population-based cohort were analyzed with regard to equol production, based on serum equol concentrations in fasting blood samples. We also compared the demographic anthropometric characteristics and blood biochemistry phenotypes between equol producers and nonproducers. We identified host genetic factors of the equol-producing phenotype using high-throughput SNP chip data and analyzed their association with host defense, providing evidence of the interaction between genetic factors and equol phenotypes, which contributes to health-related outcomes, such as blood pressure.
Materials and methods
Study participants
Study subjects were selected from an ongoing population-based cohort, as part of the Korean Genome and Epidemiology Study (KoGES). Participants were recruited from residents in two cities (Ansung and Ansan) in Gyeonggi-do province, Korea. We enrolled 10,038 men and women from 2001 to 2002 for a baseline study, whose demographics have been reported (Ko et al. 2011). From this cohort, we selected 1391 healthy adults who had never been diagnosed with any chronic disease, including cancer, diabetes, and hypertension, at baseline enrollment to measure serum equol levels. The study was approved by the Institutional Review Board of the Korea National Institute of Health, and all participants provided informed consent for participation.
All participants were questioned by trained interviewers, and general information including age, gender, smoking habits, and alcohol consumption was collected through a questionnaire. In addition, the participants’ anthropometric characteristics (blood pressure, weight, height, hip circumference, waist circumference) were measured. Ten-milliliter samples of fasting venous blood was collected in a plain tube. The serum was separated by centrifugation at 3000 rpm for 10 min at 4 °C and stored in liquid nitrogen. Biochemical parameters were measured, including fasting serum glucose, fasting serum insulin, total cholesterol, triglyceride, and HDL cholesterol.
Serum equol measurement
Equol was measured by NEODIN Medical Institute inc., Seoul, Korea, by liquid chromatography/tandem mass spectrometry (LC-MS/MS). The samples were prepared, and equol was measured per Grace et al. (2003). There was a patent binomial distribution in equol concentrations (Supplementary Figure 1), and the estimated limit of detection (LOD) was 0.068 μg/L. Thus, equol producers and nonproducers were distinguished based on the LOD, as established in a study of Korean subjects (Akaza et al. 2004).
Genotypes
Genomic DNA was extracted from the peripheral blood of participants using RBC, cell, and protein lysis solutions (INTRON biotechnology, Gyunggi-do, Korea) per the manufacturer’s instructions. DNA concentration and purity were measured on a NanoDrop spectrophotometer (Thermo, Wilmington, DE), and the integrity of genomic DNA was assessed by agarose gel electrophoresis. DNA was genotyped using the affymetrix genomewide human SNP array 5.0 at final concentrations of 100 μg/ml and 500 ng (Affymetrix, Inc., Santa Clara, CA). The quality control steps have been described (Cho et al. 2009; Rabbee and Speed 2006). Briefly, SNPs with a missing genotype call rate >0.1, minor allele frequency <0.01, and Hardy–Weinberg equilibrium (HWE) (p < 1 × 10−6) were excluded; ultimately, we used 333,651 SNPs that were genotyped in the Korean association resource (KARE) study (Cho et al. 2009).
Statistical analysis
The demographic, anthropometric, and blood biochemical properties between equol producers and nonproducers were compared by χ2 and t tests. Mean differences between producers and nonproducers were analyzed by multiple linear regression, controlling for area, sex, age, and smoking and alcohol consumption. The GWAS on equol producers and nonproducers was performed by logistic regression, controlling for covariates such as area, age, and sex. SNPs that had a p value <1 × 10−5 were selected, and the interactions between genotype and the equol-producing phenotype were examined with regard to anthropometric and blood biochemical parameters. A general linear analysis model with type III sum of squares was used to test the interactions.
Statistical analyses were performed using PLINK, version 1.07, using default options (Purcell et al. 2007), and SAS version 9.0. Trait-associated SNPs were analyzed with regard to transcription factor binding sites (TFBS) using TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html).
Results
Comparison of characteristics between equol producers and nonproducers
Serum equol concentrations in the 1391 subjects ranged from 0.03 to 3548.26 μg/L. The estimated LOD was 0.068 μg/L, below which patients were considered equol nonproducers. Thus, there were 416 equol nonproducers and 975 equol producers (70.1 %).
Our subjects came from two community-based cohorts (Ansung and Ansan); the rate of equol producers was higher in Ansan (75 %) than in Ansung (57 %). The mean age of equol producers (51.6 ± 8.4 years) was lower than that of nonproducers (52.9 ± 8.3 years). Seventy-five percent of men were equol producers versus 65 % of women. In addition, the rates of equol producers were higher in current drinkers (73 %) and current smokers (72 %) than in those who never drank (66 %) or smoked (67 %). Table 1 shows our analysis of area, age, gender, smoking, and alcohol consumption as covariates.
The anthropometric, glucose, and lipid indices are shown in Table 1. The equol-producing phenotype was significantly associated with lower pulse, systolic and diastolic blood pressure, and greater height by student t test (two-tailed p values <0.05) compared with the equol nonproducer phenotype. By linear regression, diastolic blood pressure correlated significantly with equol production (beta ± SE = −1.35 ± 0.67, p value = 0.045), after adjusting for area, age, and sex. Further analysis, adjusting for smoking and alcohol consumption, showed that these factors had no influence on the association between diastolic blood pressure and the equol-producing phenotype. The significant correlation between the equol phenotype and height could be attributed to the gender or age distribution in the study population, because the significance declined by linear regression after adjusting for age and sex (Table 1).
Genomewide association study of the equol-producing phenotype
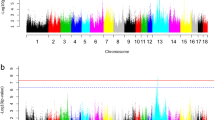
To identify genetic factors of the equol-producing phenotype, we performed a GWAS. The association p values are shown in a Manhattan plot (Supplementary Figure 1), which displays the minus log10-transformed p values on the chromosomal position of each SNP. There was no SNP that satisfied the genomewide level of significance (p value <5 × 10−8), likely due to our small sample size or the complexity of the impact of host genetic factors on the maintenance of equol-producing bacterial species.
By establishing a moderate level of significance (p value <1 × 10−5) to avoid type II errors and screening genetic variants that had small effects on the trait, we identified five signals (marked by circles in Fig. 1 and listed in Table 2), all of which were clustered in the 6q21 region, encompassing an E3 ubiquitin-protein ligase-coding gene (HACE1) (Fig. 2). The r 2 value of the five SNPs exceeded 0.95, indicating that they were in strong linkage disequilibrium (LD), excluding the possibility of sporadic association. The most significant SNP was rs6927608A > C, and individuals with this minor allele were equol nonproducers (OR = 0.57, CI 0.45–0.72; p value = 2.5 × 10−6), suggesting that individuals with the minor allele are unable to produce equol from daidzein, possibly due to the HACE1-mediated absence of equol-producing bacteria in their intestinal microflora.
We surveyed transcription factor binding sites around the SNPs, as shown in Table 2. Although the most significant SNP (rs6927608) did not lie in a transcription factor binding site (TFBS), one of the significant SNPs (rs17065302C > G, Table 2), strongly linked to rs6927608, was found in an activator protein 1 (AP-1) motif, which is implicated in estrogen receptor-mediated regulation of gene expression. This finding suggests a connection between HACE1 and the equol-producing phenotype, based on the structural similarity of equol to the potent estrogen estradiol (Setchell and Clerici 2010 as a review). In conclusion, our GWAS identified host genetic factors that involve HACE1 polymorphisms in the equol-producing phenotype.
Interactive effect of the rs6927608 genotype and the equol-producing phenotype on blood pressure
To determine whether the association between the HACE1 polymorphisms and interindividual differences in the equol-producing phenotype impacted other phenotypic outcomes, rs6927608 was analyzed with regard to whether the interaction between equol and SNPs had cumulative effects on clinical indices. A general linear model was applied to test the interaction, and the results are shown in Table 3. Notably, there was a statistically significant effect of the equol × SNP interaction on systolic blood pressure (p value = 1.3 × 10−4)—more significant than with equol alone.
Based on the mean blood pressure in equol producers and nonproducers at each genotype, as shown in Fig. 2, equol producers who were major allele homozygotes (AA) had lower systolic blood pressure than equol nonproducers, but the presence of the minor allele (C) in equol producers progressively increased blood pressure (beta ± SE = 3.6 ± 1.1, p = 0.0015). However, the genotype effect on blood pressure in equol nonproducers was opposite to that of equol producers, demonstrating that the minor allele (C) in equol nonproducers is linked to decreased systolic blood pressure (beta ± SE = −3.5 ± 1.5, p = 0.017).
Discussion
The most pronounced effect of the equol-producing phenotype is its ability to lower blood pressure. Equol producers had significantly decreased blood pressure than nonproducers by t test and linear regression analysis, after controlling for area, age, and sex. The relationship between equol production and blood pressure was reported by Tormala et al. (2007), who examined blood pressure in tibolone-treated postmenopausal women and concluded that equol producers had lower blood pressure compared with nonproducers. Additional reports have suggested that soy products reduce the risk of coronary artery disease (Clarkson 2002; Messina et al. 2002).
Our GWAS identified a candidate gene as a host genetic factor, HACE1, which encodes C-terminal homologous to E6-associating protein carboxyl terminus (HECTc) ubiquitin-protein ligase domain and ankyrin repeat-containing E3 ubiquitin-protein ligase 1. HACE1 lies on chromosome 6q21, and translocations in this region have been reported in Wilms’ tumor (Bruce et al. 2003). The human gene is expressed in many tissues, including heart, brain, placenta, kidney, and pancreas (Anglesio et al. 2004)—predominantly in the endoplasmic reticulum membrane (Zhang et al. 2007). HACE1 is a tumor suppressor, and genetic inactivation of HACE1 in mice causes the development of spontaneous, late-onset cancer (Zhang et al. 2007). In spite of these previous findings, however, no relationship between HACE1 function, equol phenotype, and blood pressure has been reported.
Our analysis revealed that the equol × HACE1 SNP (rs6927608) interaction correlated robustly with systolic blood pressure (SBP, p value = 1.3 × 10−4 in Table 3), showing a more significant association in the interactive model compared with the association between SBP and equol. Notably, rs6927608 was not associated with blood pressure by itself. Lowered blood pressure was observed only in homozygous carriers of the major allele of rs6927608. Although a GWAS has identified common variants that are reproducibly associated with blood pressure and the risk of stroke and ischemic heart disease, such variants explain merely 1 % of the variation in blood pressure (Levy et al. 2009), which indicates a multifactorial nature of the regulation of blood pressure and the etiology of related diseases. Our findings on the interactive (or cumulative) effects of a host’s genetic and environmental factors on blood pressure reflect the complexity of the trait and increase our understanding of the mechanism by which blood pressure is regulated.
One possible mechanism of the association of HACE1 with equol–producing phenotype is its involvement in host immune responses. Notably, a GWAS from the Framingham Heart Study showed that HACE1 correlates with serum thyroid-stimulating hormone (TSH) concentrations (Hwang et al. 2007). Further, HACE1 is upregulated in natural killer T-cell lymphomas Huang et al. (2010). Considering that TSH and natural killer T cells mediate the immune response (Wang and Klein 2001), HACE1 appears to participate in host defense by mediating the TSH and natural killer T-cell functions.
HACE1 expression rises in the mesenchymal fraction of embryonic small intestine versus the epithelial fraction, as shown in an expression profile analysis from a gene expression omnibus database (GDS2699, http://www.ncbi.nlm.gov/geoprofiles, Supplementary Figure 1). Thus, we implicate HACE1 in intestinal immune responses, which might influence the regulation and maintenance of the microbial environment in the host gut.
The search for TFBS suggested that the equol-producing trait-associated HACE1 polymorphisms might have functional implication in the HACE1 expression. We noted that one such SNP, rs17065302C > G, lay in a possible AP-1 binding site (TFBS homology to the consensus sequence: 93.5 %), with the major allele “ATGACTCA” (Carroll et al. 2006). The replacement of cytosine with guanine in the minor allele of rs17065302 eliminates the AP-1 motif (ATGACTGA)—that is, carriers of the major allele who express the equol-producing phenotype harbor the AP-1 binding site in HACE1.
The AP-1 transcription factor family drives estrogen receptor (ER)-mediated gene transcription by binding to AP-1 response elements in target gene promoters in response to the binding of ER to its ligand, such as estradiol (Jakacka et al. 2001). Equol has a similar structure to estradiol and has high affinity for ER, acting as its ligand (Setchell and Clerici 2010). Based on these data, we speculate that HACE1 expression in individuals with the major allele (harboring AP-1 sites) is altered by the equol–ER complex that is bound to AP-1 sites. Considering the involvement of HACE1 in the immune system, as discussed above, equol-induced modulations in HACE1 expression might affect host intestinal immune responses, which might in turn render an intestinal environment favorable for maintaining equol-producing bacteria. Nevertheless, functional implication of HACE1 polymorphisms in determining phenotypic outcomes is unknown, and alternative models for the functions of other associated polymorphisms should be considered.
Our study demonstrates that equol production is linked to blood pressure and that HACE1 is a determinant of the equol-producing phenotype, supported by LC-MS/MS-based quantitative measurements of serum equol concentrations and our GWAS analysis. Although further studies should be performed to confirm the association in other populations and investigate the underlying mechanism, we propose that a host’s genetic predisposition and physiological environment cumulatively influence disease-related phenotypic outcomes, such as blood pressure. Based on the complex interaction between a host and its physiological microbial environment, as indicated by recent meta-genomic data, an individual’s genetic background is believed to influence the intestinal bacteria profile.
We believe that our findings provide significant insight into the functions of host factors that govern the capacity to harbor specific intestinal microenvironments, including daidzein-metabolizing bacteria, which in turn has implications in a host’s health-related phenotypic outcomes. Further, these results should contribute to the development of well-designed clinical studies that will allow us to substantiate and better understand the effects of equol and isoflavone on health.
References
Akaza H, Miyanaga N, Takashima N, Naito S, Hirao Y, Tsukamoto T, Mori M (2002) Is daidzein non-metabolizer a high risk for prostate cancer? A case-controlled study of serum soybean isoflavone concentration. Jpn J Clin Oncol 32:296–300
Akaza H, Miyanaga N, Takashima N, Nagata Y, Hirao Y, Tsukamoto T, Fujioka T, Mori M, Kim WJ, Song JM, Pantuck AJ (2004) Comparisons of percent equol producers between prostate cancer patients and controls: case-controlled studies of isoflavones in Japanese, Korean and American residents. Jpn J Clin Oncol 34:86–89
Alekel DL, Van Loan MD, Koehler KJ, Hanson LN, Stewart JW, Hanson KB, Kurzer MS, Peterson CT (2010) The soy isoflavones for reducing bone loss (SIRBL) study: a 3-y randomized controlled trial in postmenopausal women. Am J Clin Nutr 91:218–230
Andres S, Abraham K, Appel KE, Lampen A (2011) Risks and benefits of dietary isoflavones for cancer. Crit Rev Toxicol 41:463–506
Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, Leach S, Marra MA, Brooks-Wilson AR, Penninger J, Sorensen PHB (2004) Differential expression of a novel arkyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Hum Mol Genet 13:2061–2074
Atkinson C, Berman S, Humbert O, Lampe JW (2004a) In vitro incubation of human feces with daidzein and antibiotics suggests interindividual differences in the bacteria responsible for equol production. J Nutr 134:596–599
Atkinson C, Berman S, Humbert O, Lampe JW (2004b) The in vitro metabolism of daidzein by human intestinal bacteria. J Nutr 134:596–599
Atkinson C, Frankenfeld CL, Lampe JW (2005) Gut bacterial metabolism of the soy isoflavone daidzein: exploring the relevance to human health. Exp Biol Med 230:155–170
Brink E, Coxam V, Robins S, Wahala K, Cassidy A, Branca F (2008) Long-term consumption of isoflavone-enriched foods does not affect bone mineral density, bone metabolism, or hormonal status in early postmenopausal women: a randomized, double-blind, placebo controlled study. Am J Clin Nutr 87:761–770
Bruce CK, Howard P, Nowak NJ, Hoban PR (2003) Molecular analysis of region t(5;6)(q21;q21) in Wilms tumor. Cancer Genet Cytogenet 141:106–113
Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton RK, Fertuck KC, Hall GE, Wang Q, Berkiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genomewide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297
Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL (2009) A large-scale genomewide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 41:527–534
Clarkson TB (2002) Soy, soy phytoestrogens and cardiovascular disease. J Nutr 132:566S–569S
Dai Q, Franke AA, Jin F, Shu XO, Hebert JR, Custer LJ, Cheng J, Gao YT, Zheng W (2002) Urinary excretion of phytoestrogens and risk of breast cancer among Chinese women in Shanghai. Cancer Epidemiol Biomarkers Prev 11:815–821
DellaPenna D (1999) Nutritional genomics: manipulating plant micronutrients to improve human health. Science 285:375–379
Grace PB, Taylor JI, Botting NP, Fryatt T, Oldfield MF, Al-Maharik N, Bingham SA (2003) Quantification of isoflavones and lignans in serum using isotope dilution liquid chromatography tandem mass spectrometry. Rapid Commun Mass Spectrom 17:1350–1357
Heinonen S, Wähälä K, Adlercreutz H (1999) Identification of isoflavone metabolites dihydrodaidzein, dihydrogenistein, 6-OH-O-dma, and cis-4-OH-equol in human urine by gas chromatography-mass spectroscopy using authentic reference compounds. Anal Biochem 274:211–219
Huang Y, de Reyniès A, de Leval L, Ghazi B, Martin-Garcia N, Travert M, Bosq J, Brière J, Thomas E, Coppo P, Marafioti T, Emile JF, Delfau-Larue MH, Schmitt C, Gaulard P (2010) Gene expression profiling identifies emerging oncogenic pathway operating in extranodal NK/T-cell lymphoma, nasal type. Blood 115:1226–1236
Hwang SJ, Yang Q, Meigs JB, Pearce EN, Fox CS (2007) A genomewide association for kidney function and endocrine-related traits in the NHLBI’s Framingham heart study. BMC Med Genet 8:S10
Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL (2001) Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem 276:13615–13621
Ko KP, Min H, Ahn Y, Park SJ, Kim CS, Park JK, Kim SS (2011) A prospective study investigating the association between environmental tobacco smoke exposure and the incidence of type 2 diabetes in never smokers. Ann Epidemiol 21:42–47
Kwon DY, Daily JW III, Kim HJ, Park S (2010) Antidiabetic effects of fermented soybean products on type 2 diabetes. Nutr Res 30:1–13
Levy D, Ehret GB, Rice K et al (2009) Genomewide association study of blood pressure and hypertension. Nat Genet 41:677–687
Ma DF, Qin LQ, Wang PY, Katoh R (2008) Soy isoflavone intake increases bone mineral density in the spine of menopausal women: meta-analysis of randomized controlled trials. Clin Nutr 27:57–64
Messina M, Gardner C, Barnes S (2002) Gaining insight into the health effects of soy but a long way still to go: commentary on the fourth international symposium on the role of soy in preventing and treating chronic disease. J Nutr 132:547S–551S
O’Hara AM, Shanahan F (2006) The gut flora as a forgotten organ. EMBO Rep 7:688–693
Ozasa K, Nakao M, Watanabe Y, Hayashi K, Miki T, Mikami K, Mori M, Sakauchi F, Washio M, Ito Y, Suzuki K, Wakai K, Group AT (2004) Serum phytoestrogens and prostate cancer risk in a nested case-control study among Japanese men. Cancer Sci 95:65–71
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Rabbee N, Speed TP (2006) A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics 22:7–12
Rowland I, Berman S, Humber O, Lampe JW (1999) Metabolism of oestrogen and phytoestrogens: role of the gut microflora. Biochem Soc Trans 27:304–308
Sakar FH, Li YW (2002) Mechanisms of cancer chemoprevention by soy isoflavone genistein. Cancer Metast Rev 21:265–280
Setchell KDR (1998) Phytoestrogens: the biochemistry, physiology, and implications for human health of soy isoflavones. Am J Clin Nutr 68:1333S–1346S
Setchell KDR, Clerici C (2010) Equol: parmacokinetics and biological actions. J Nutr 140:1363S–1368S
Setchell KDR, Brown NM, Lydeking-Olsen E (2002) The clinical importance of the metabolite equol-a clue to the effectiveness of soy and its isoflavones. J Nutr 132:3577–3584
Song KB, Atkinson C, Frankenfeld CL, Jokela T, Wähälä K, Tomas WK, Lampe JW (2006) Prevalence of daidzein-metabolizing phenotypes differs between Caucasian and Korean American women and girls. J Nutr 136:1347–1351
Tormala RM, Appt S, Clarkson TB, Tikkanen MJ, Ylikorkala O, Mikkola TS (2007) Individual differences in equol production capability modulate blood pressure in tibolone-treated postmenopausal women: lack of effect of soy supplementation. Climacteric 10:471–479
Wang HC, Klein JR (2001) Immune function of thyroid stimulating hormone and receptor. Crit Rev Immunol 21:327–337
Yuan JP, Wang JH, Liu X (2007) Metabolism of dietary soy isoflavones to equol by human intestinal microflora-implications for health. Mol Nutr Food Res 51:765–781
Zhang L, Anglesio M, O’Sullivan M, Zhang F, Yang G, Sarao R, Nghiem MP, Farrar J, Arceci RJ, Sorensen PH, Penninger JM (2007) The E3 ligase HACE1 is a critical chromosomal 6q21 tumor suppressor involved in multiple cancers. Nat Med 13:1060–1069
Acknowledgments
This work was supported by the Korean Genome Analysis Project (4845-301) and the KoGES (4851-302), funded by the Ministry for Health and Welfare, Republic of Korea.
Conflict of interest
There is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12263_2012_292_MOESM1_ESM.tif
Supplementary Figure 1 Manhattan plot: minus log10-transformed p values on the chromosomal position of the respective SNP (dot). The black circles highlight the 5 most significant SNPs. (TIFF 54 kb)
12263_2012_292_MOESM2_ESM.tif
Supplementary Figure 2 Gene expression profiles obtained from GEO database (www.ncbi.nlm.nih.gov/geo-profiles). HACE1 expression in intestinal epithelium and mesenchyme of mouse embryonic small intestine. (TIFF 261 kb)
Rights and permissions
About this article
Cite this article
Hong, KW., Ko, KP., Ahn, Y. et al. Epidemiological profiles between equol producers and nonproducers: a genomewide association study of the equol-producing phenotype. Genes Nutr 7, 567–574 (2012). https://doi.org/10.1007/s12263-012-0292-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12263-012-0292-8