Abstract
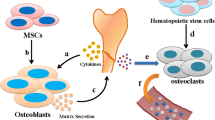
Mesenchymal stem cells (MSCs) are an important population of multipotent stem cells that differentiate into multiple lineages and display great potential in bone regeneration and repair. Although the role of protein-coding genes in the osteogenic differentiation of MSCs has been extensively studied, the functions of noncoding RNAs in the osteogenic differentiation of MSCs are unclear. The recent application of next-generation sequencing to MSC transcriptomes has revealed that long noncoding RNAs (lncRNAs) are associated with the osteogenic differentiation of MSCs. LncRNAs are a class of non-coding transcripts of more than 200 nucleotides in length. Noncoding RNAs are thought to play a key role in osteoblast differentiation through various regulatory mechanisms including chromatin modification, transcription factor binding, competent endogenous mechanism, and other post-transcriptional mechanisms. Here, we review the roles of lncRNAs in the osteogenic differentiation of bone marrow- and adipose-derived stem cells and provide a theoretical foundation for future research.



Similar content being viewed by others
References
Djebali, S., Davis, C. A., Merkel, A., et al. (2012). Landscape of transcription in human cells. Nature, 489(7414), 101–108.
Esteller, M. (2011). Non-coding RNAs in human disease. Nature Reviews Genetics, 12(12), 861 – 74.
Fu, X. D. (2014). Non-coding RNA: a new frontier in regulatory biology. National Science Review, 1, 190–204.
Sone, M., Hayashi, T., Tarui, H., et al. (2007). The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. Journal of Cell Science, 120(Pt 15), 2498 – 506.
Guttman, M., Donaghey, J., Carey, B. W., et al. (2011). LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature, 477(7364), 295–300.
Wapinski, O., & Chang, H. Y. (2011). Long noncoding RNAs and human disease. Trends in Cell Biology, 21(6), 354 – 61.
Guttman, M., & Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature, 482(7385), 339 – 46.
Ma, L., Bajic, V. B., & Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biology, 10(6), 925 – 33.
Cabili, M. N., Trapnell, C., Goff, L., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development, 25(18), 1915–1927.
Ulitsky, I., & Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell, 154(1), 26–46.
Bartolomei, M. S., Zemel, S., & Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature, 351(6322), 153–155.
Willingham, A. T., Orth, A. P., Batalov, S., et al. (2005). A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science, 309(5740), 1570–1573.
Rinn, J. L., Kertesz, M., Wang, J. K., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 129(7), 1311–1323.
Martianov, I., Ramadass, A., Serra Barros, A., Chow, N., & Akoulitchev, A. (2007). Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature, 445(7128), 666 – 70.
Lee, J. T. (2009). Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes & Development, 23(16), 1831–1842.
Lv, D., Sun, R., Yu, Q., & Zhang, X. (2016). The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer. Tumour Biology.
Rumman, M., Dhawan, J., & Kassem, M. (2015). Concise review: quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cells, 33, 2903–2912.
Bianco, P., Riminucci, M., Gronthos, S., & Robey, P. G. (2001) Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells, 19, pp. 180 – 92.
Pittenger, M. F., Mackay, A. M., Beck, S. C., et al. (1999). Multilineage potential of adult human mesenchymal stem cells. Science, 284(5411), 143–147.
da Silva Meirelles, L., Chagastelles, P. C., & Nardi, N. B. (2006). Mesenchymal stem cells reside in virtually all post-natal organs and tissues. Journal of Cell Science, 119, 2204–2213.
Quarto, R., Mastrogiacomo, M., Cancedda, R., et al. (2001). Repair of large bone defects with the use of autologous bone marrow stromal cells. The New England Journal of Medicine, 344(5), 385–386.
Rastegar, F., Rastegar, F., Shenaq, D., et al. (2010). Mesenchymal stem cells: molecular characteristics and clinical applications. World Journal of Stem Cells, 2(4), 67–80.
Gaur, T., Lengner, C. J., Hovhannisyan, H., et al. (2005). Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. The Journal of Biological Chemistry, 280(39), 33132–33140.
Chen, G., Deng, C., & Li, Y. P. (2012). TGF-beta and BMP signaling in osteoblast differentiation and bone formation. International Journal of Biological Sciences, 8(2), 272 – 88.
Deng, Z. L., Sharff, K. A., Tang, N., et al. (2008). Regulation of osteogenic differentiation during skeletal development. Frontiers in Bioscience, 13, 2001–2021.
Javed, A., Bae, J. S., Afzal, F., et al. (2008). Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. The Journal of Biological Chemistry, 283(13), 8412–8422.
Hong, J. H., & Yaffe, M. B. (2006). TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle, 5(2), 176–179.
Chen, Q., Shou, P., Zheng, C., et al. (2016). Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death and Differentiation, 23(7), 1128–1139.
Li, H., Li, T., Fan, J., et al. (2015). miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death and Differentiation, 22(12), 1935–1945.
Zhang, J. F., Fu, W. M., He, M. L., et al. (2011). MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biology, 8(5), 829 – 38.
Huang, S., Wang, S., Bian, C., et al. (2012). Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells and Development, 21(13), 2531–2540.
Wang, J., Guan, X., Guo, F., et al. (2013). miR-30e reciprocally regulates the differentiation of adipocytes and osteoblasts by directly targeting low-density lipoprotein receptor-related protein 6. Cell Death & Disease, 4, e845.
Li, Z., Hassan, M. Q., Volinia, S., et al. (2008). A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proceedings of the National Academy of Sciences of the United States of America, 105(37), 13906–13911.
Zhang, W., Dong, R., Diao, S., Du, J., Fan, Z., & Wang, F. (2017). Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Research & Therapy, 8, 30.
Tye, C. E., Gordon, J. A., Martin-Buley, L. A., Stein, J. L., Lian, J. B., & Stein, G. S. (2015). Could lncRNAs be the missing links in control of mesenchymal stem cell differentiation? Journal of Cellular Physiology, 230(3), 526 – 34.
Wang, L., Wang, Y., Li, Z., & Yu, B. (2015). Differential expression of long noncoding ribonucleic acids during osteogenic differentiation of human bone marrow mesenchymal stem cells. International Orthopaedics, 39(5), 1013–1019.
Zuo, C., Wang, Z., Lu, H., Dai, Z., Liu, X., & Cui, L. (2013). Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Molecular Medicine Reports, 8(2), 463–467.
Song, W. Q., Gu, W. Q., Qian, Y. B., Ma, X., Mao, Y. J., & Liu, W. J. (2015). Identification of long non-coding RNA involved in osteogenic differentiation from mesenchymal stem cells using RNA-Seq data. Genetics and Molecular Research, 14(4), 18268–18279.
Cui, Y., Lu, S., Tan, H., Li, J., Zhu, M., & Xu, Y. (2016). Silencing of long non-coding RNA NONHSAT009968 ameliorates the staphylococcal protein a-inhibited osteogenic differentiation in human bone mesenchymal stem cells. Cellular Physiology and Biochemistry, 39(4), 1347–1359.
Cai, X., & Cullen, B. R. (2007). The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA, 13(3), 313–316.
Huang, Y., Zheng, Y., Jia, L., & Li, W. (2015). Long noncoding RNA H19 promotes osteoblast differentiation via TGF-beta1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells, 33(12), 3481–3492.
Liang, W. C., Fu, W. M., Wang, Y. B., et al. (2016). H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Scientific Reports, 6, 20121.
Ohnishi, Y., Tanaka, T., Yamada, R., et al. (2000). Identification of 187 single nucleotide polymorphisms (SNPs) among 41 candidate genes for ischemic heart disease in the Japanese population. Human Genetics 106(3), 288 – 92.
Vausort, M., Wagner, D. R., & Devaux, Y. (2014). Long noncoding RNAs in patients with acute myocardial infarction. Circulation Research, 115(7), 668 – 77.
Yan, B., Yao, J., Liu, J. Y., et al. (2015). lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circulation Research, 116(7), 1143–1156.
Barry, G., Briggs, J. A., Vanichkina, D. P., et al. (2014). The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular Psychiatry, 19(4), 486 – 94.
Jin, C., Zheng, Y., Huang, Y., Liu, Y., Jia, L., Zhou, Y., et al. (2017). Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells. Cell Biology International, 41(1), 33–41.
Shen, Y., Dong, L. F., Zhou, R. M., et al. (2016). Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. Journal of Cellular and Molecular Medicine, 20(3), 537 – 48.
Kretz, M., Webster, D. E., Flockhart, R. J., et al. (2012). Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes & Development, 26(4), 338 – 43.
Zhu, L., & Xu, P. C. (2013). Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochemical and Biophysical Research Communications, 432(4), 612–617.
Jia, Q., Jiang, W., & Ni, L. (2015). Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Archives of Oral Biology, 60(2), 234 – 41.
Jia, Q., Chen, X., Jiang, W., Wang, W., Guo, B., & Ni, L. (2016). The Regulatory Effects of Long Noncoding RNA-ANCR on Dental Tissue-Derived Stem Cells. Stem Cells International, 2016, 3146805.
Chisholm, K. M., Wan, Y., Li, R., Montgomery, K. D., Chang, H. Y., & West, R. B. (2012). Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One, 7(10), e47998.
Endo, H., Shiroki, T., Nakagawa, T., et al. (2013). Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One, 8(10), e77070.
Yang, Z., Zhou, L., Wu, L. M., et al. (2011). Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Annals of Surgical Oncology, 18(5), 1243–1250.
Xing, D., Liang, J. Q., Li, Y., et al. (2014). Identification of long noncoding RNA associated with osteoarthritis in humans. Orthopaedic Surgery, 6(4), 288 – 93.
Wei, B., Wei, W., Zhao, B., Guo, X., & Liu, S. (2017). Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of femoral head. PLoS One, 12(2), e0169097.
Anwar, S. L., Krech, T., Hasemeier, B., et al. (2012). Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PloS One, 7, e49462.
Zhou, Y., Zhang, X., & Klibanski, A. (2012). MEG3 noncoding RNA: a tumor suppressor. Journal of Molecular Endocrinology, 48(3), R45-53.
Zhuang, W., Ge, X., Yang, S., et al. (2015). Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells, 33(6), 1985–1997.
Li, Z., Jin, C., Chen, S., et al. (2017). Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Molecular and Cellular Biochemistry.
Wang, Q., Li, Y., Zhang, Y., et al. (2017). LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother, 89, 1178–1186.
Zhao, Y., Feng, G., Wang, Y., Yue, Y., & Zhao, W. (2014). Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis. International Journal of Clinical and Experimental Pathology, 7(11), 7643–7652.
Wang, S., Zhang, X., Yuan, Y., et al. (2015). BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. European Journal of Cardio-Thoracic Surgery, 47(3), 439–46.
Xu, Y., Wang, S., Tang, C., & Chen, W. (2015). Upregulation of long non-coding RNA HIF 1alpha-anti-sense 1 induced by transforming growth factor-beta-mediated targeting of sirtuin 1 promotes osteoblastic differentiation of human bone marrow stromal cells. Molecular Medicine Reports, 12(5), 7233–7238.
Wang, Y., Dang, Y., Liu, J., & Ouyang, X. (2016). The function of homeobox genes and lncRNAs in cancer. Oncology Letters, 12(3), 1635–1641.
Zhu, X. X., Yan, Y. W., Chen, D., et al. (2016). Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget, 7(39), 63561–63570.
Augoff, K., McCue, B., Plow, E. F., & Sossey-Alaoui, K. (2012). miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Molecular Cancer, 11, 5.
Yang, H., Liu, P., Zhang, J., et al. (2016). Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene, 35(28), 3647–3657.
Jin, C., Jia, L., Huang, Y. et al. (2016). Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose-derived stem cells. Stem Cells.
Sheik Mohamed, J., Gaughwin, P. M., Lim, B., Robson, P., & Lipovich, L. (2010). Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA, 16(2), 324 – 37.
Li, H., Li, H., Zhang, Z., Chen, Z., & Zhang, D. (2015). Osteogenic growth peptide promotes osteogenic differentiation of mesenchymal stem cells mediated by LncRNA AK141205-induced upregulation of CXCL13. Biochemical and Biophysical Research Communications, 466(1), 82 – 8.
Cao, B., Liu, N., & Wang, W. (2016). High glucose prevents osteogenic differentiation of mesenchymal stem cells via lncRNA AK028326/CXCL13 pathway. Biomedicine and Pharmacotherapy, 84, 544–551.
Pasquinelli, A. E. (2012). MicroRNAs and their targets: recognition, regu- lation and an emerging reciprocal relationship. Nature Reviews Genetics, 13, 271–282.
Quinn, J. J., & Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics, 17, 47–62.
Egger, G., et al. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature, 429(6990), 457 –63.
Helin, K., & Dhanak, D. (2013). Chromatin proteins and modifications as drug targets. Nature, 502(7472), 480–488.
Hemming, S., Cakouros, D., senmann, S., et al. (2014). EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells, 32(3), 802–15.
Guil, S., Soler, M., Portela, A., et al. (2012). Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nature Structural and Molecular Biology, 19(7), 664–70.
Ghosal, S., Das, S., & Chakrabarti, J. (2013). Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells and Development, 22(16), 2240–2253.
Khalil, A. M., Guttman, M., Huarte, M., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America, 106(28), 11667–11672.
Miyoshi, N., Wagatsuma, H., Wakana, S., et al. (2000). Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes to Cells, 5(3), 211–20.
Tay, Y., Rinn, J., & Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature, 505(7483), 344–52.
Xing, C. Y., Hu, X. Q., Xie, F. Y., et al. (2015). Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Letters, 589(15), 1981–1987.
Cai, H., Xue, Y., Wang, P., et al. (2015). The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget, 6(23), 19759–19779.
Keniry, A., et al. (2012). The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature Cell Biology, 14(7), 659–65.
Wu, Y., et al. (2014). Long noncoding RNA HOTAIR involvement in cancer. Tumour Biology, 35(10), 9531–9538.
Kogure, T., Yan, I. K., Lin, W. L., & Patel, T. (2013). Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer, 4(7–8), 261–72.
Conigliaro, A., Costa, V., Lo Dico, A., et al. (2015). CD90 + liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Molecular Cancer, 14, 155.
Thery, C., Amigorena, S., Raposo, G., & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, 3, Unit 3 22.
Hewson, C., & Morris, K. V. (2016). Form and Function of Exosome-Associated Long Non-coding RNAs in Cancer. Current Topics in Microbiology and Immunology, 394, 41–56.
Acknowledgements
This study was financially supported by grants from the National Natural Science Foundation of China (Nos. 81670957, 81772876, and 81700938), a grant from the Peking University (PKU) School and Hospital of Stomatology (No. PKUSS20140104), and seed grants from the PKU School of Stomatology for PostDoc (No. YS0203) and the Beijing Natural Science Foundation (No. 7172239).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest to declare.
Rights and permissions
About this article
Cite this article
Yang, Q., Jia, L., Li, X. et al. Long Noncoding RNAs: New Players in the Osteogenic Differentiation of Bone Marrow- and Adipose-Derived Mesenchymal Stem Cells. Stem Cell Rev and Rep 14, 297–308 (2018). https://doi.org/10.1007/s12015-018-9801-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-018-9801-5