Abstract
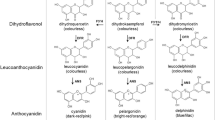
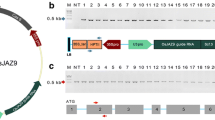
Altering a trait by CRISPR-Cas9-targeted mutagenesis offers great advantages in identifying gene function and crop improvement. In the present study, three genes (OsF3′H, OsDFR and OsLDOX) in the anthocyanin biosynthesis pathway were successfully edited on the Heugseonchal or Sinmyungheugchal variety using the CRISPR/Cas9 system. As a result, the ratio of the edited plants in the transformed early generation was 56.7%. These edited mutant lines were observed with the changes of seed color and anthocyanin content. All mutations were stably inherited to the T2 progeny. In addition, we could select edited homozygous mutant lines lacking the T-DNA already in the first offspring generation. Also the insertion of vector backbone sequences in f3′h-9, dfr-4 and ldox-16 lines was not detected in the whole genome resequencing. These results demonstrated that the CRISPR/Cas9 system can induce clearly gene-specific mutations with a high efficiency in rice and null plants selected from these mutants cannot be distinguished from non-GMO plants even under strict GMO regulation.





Similar content being viewed by others
References
Abdel-Aal ES, Hucl P (1999) A rapid method for quantifying total anthocyanins in blue aleurone and purple pericarp wheats. Cereal Chem 76:350–354. https://doi.org/10.1094/CCHEM.1999.76.3.350
Abdel-Aal ESM, Hucl P (2003) Composition and stability of anthocyanins in blue-grained wheat. J Agr Food Chem 51:2174–2180. https://doi.org/10.1021/jf021043x
Abdel-Aal ESM, Young JC, Rabalski I (2006) Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J Agr Food Chem 54:4696–4704. https://doi.org/10.1021/jf0606609
Batschauer A, Rocholl M, Kaiser T, Nagatani A, Furuya M, Schäfer E (1996) Blue and UV-A light-regulated CHS expression in Arabidopsis independent of phytochrome A and phytochrome B. Plant J 9:63–69. https://doi.org/10.1046/j.1365-313X.1996.09010063.x
Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96:12929–12934. https://doi.org/10.1073/pnas.96.22.12929
Chen X, Tao Y, Al A, Zhuang Z, Guo D, Guo Q, Wang J (2019) Transcriptome and proteome profiling of different colored rice reveals physiological dynamics involved in the flavonoid pathway. Int J Mol Sci 20:2463. https://doi.org/10.3390/ijms20102463
Chen XQ, Nagao N, Itani T, Irifune K (2012) Anti-oxidative analysis, and identification and quantification of anthocyanin pigments in different coloured rice. Food Chem 135:2783–2788. https://doi.org/10.1016/j.foodchem.2012.06.098
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Furukawa T, Maekawa M, Oki T, Suda I, Iida S, Shimada H, Kadowaki KI (2007) The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J 49:91–102. https://doi.org/10.1111/j.1365-313X.2006.02958.x
Hyun JW, Chung HS (2004) Cyanidin and malvidin from Oryza sativa cv. Heugjinjubyeo mediate cytotoxicity against human monocytic leukemia cells by arrest of G2/M phase and induction of apoptosis. J Agric Food Chem 52:2213–2217. https://doi.org/10.1021/jf030370h
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat biotechnol 31:233. https://doi.org/10.1038/nbt.2508
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Kim B, Cho Y, Kim S (2017) Identification of a novel DFR-A mutant allele determining the bulb color difference between red and yellow onions (Allium cepa L.). Plant Breed Biotech 5:45–53. https://doi.org/10.1016/j.foodres.2011.07.037
Kim H, Kim ST, Ryu J, Choi MK, Kweon J, Kang BC, Kim SG (2016) A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J Integr Plant Biol 58:705–712. https://doi.org/10.1111/jipb.12474
Kim JK, Lee SY, Chu SM, Lim SH, Suh SC, Lee YT, Ha SH (2010) Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J Agric Food Chem 58:12804–12809. https://doi.org/10.1021/jf103277g
Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4:1229–1236. https://doi.org/10.1105/tpc.4.10.1229
Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23:587–596. https://doi.org/10.1046/j.1365-313x.2000.00833.x
Lee HJ, Abdula SE, Jee MG, Jang DW, Cho YG (2011) High-efficiency and Rapid Agrobacterium-mediated genetic transformation method using germinating rice seeds. J Plant Biotechnol 38:251–257. https://doi.org/10.5010/JPB.2011.38.4.251
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Plant Biol 57:405–430. https://doi.org/10.1146/annurev.arplant.57.032905.105252
Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Muday GK (2011) Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol 156:144–164. https://doi.org/10.1104/pp.111.172502
Li WX, Wu SL, Liu YH, Jin GL, Zhao HJ, Fan LJ, Shu QY (2016) Genome-wide profiling of genetic variation in Agrobacterium-transformed rice plants. J Zhejiang Univ Sci B 17:992–996. https://doi.org/10.1631/jzus.B1600301
Lim SH, Ha SH (2013) Marker development for the identification of rice seed color. Plant Biotechnol Rep 7:391–398. https://doi.org/10.1007/s11816-013-0276-1
Ma H, Pooler M, Griesbach R (2009) Anthocyanin regulatory/structural gene expression in Phalaenopsis. J Am Soc Hortic Sci 134: 88–96. https://doi.org/10.21273/JASHS.134.1.88
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Xie Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284. https://doi.org/10.1016/j.molp.2015.04.007
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. https://doi.org/10.1126/science.1232033
Mazza G, Gao L (2005) Blue and purple grains. In: Grains S, for Food and Feed. E. Abdel-Aal and P. Wood, (eds) American Association of Cereal Chemists. St. Paul, MN, pp 313–350
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217
Nakatsuka T, Abe Y, Kakizaki Y, Yamamura S, Nishihara M (2007) Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. Plant Cell Rep 26:1951–1959. https://doi.org/10.1007/s00299-007-0401-0
Nakornriab M, Sriseadka T, Wongpornchai S (2008) Quantification of carotenoid and flavonoid components in brans of some Thai black rice cultivars using supercritical fluid extraction and high-performance liquid chromatography-mass spectrometry. J Food Lipids 15:488–503. https://doi.org/10.1111/j.1745-4522.2008.00135.x
Nam SH, Choi SP, Kang MY, Koh HJ, Kozukue N, Friedman M (2006) Antioxidative activities of bran extracts from twenty one pigmented rice cultivars. Food Chem 94:613–620. https://doi.org/10.1016/j.foodchem.2004.12.010
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691. https://doi.org/10.1038/nbt.2655
Oikawa T, Maeda H, Oguchi T, Yamaguchi T, Tanabe N, Ebana K, Izawa T (2015) The birth of a black rice gene and its local spread by introgression. Plant Cell 27:2401–2414. https://doi.org/10.1105/tpc.15.00310
Park J, Lim K, Kim JS, Bae S (2017) Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33:286–288. https://doi.org/10.1093/bioinformatics/btw561
Pereira-Caro G, Watanabe S, Crozier A, Fujimura T, Yokota T, Ashihara H (2013) Phytochemical profile of a Japanese black–purple rice. Food Chem 141:2821–2827. https://doi.org/10.1016/j.foodchem.2013.05.100
Philpott M (2006) In situ and in vitro antioxidant activity of sweetpotato anthocyanins. J Agr Food Chem 54:1710–1715. https://doi.org/10.1021/jf034593j
Salinas Moreno Y, Sánchez GS, Hernández DR, Lobato NR (2005) Characterization of anthocyanin extracts from maize kernels. J Chromatogr Sci 43:483–487. https://doi.org/10.1093/chromsci/43.9.483
Schouten HJ, vande Geest H, Papadimitriou S, Bemer M, Schaart JG, Smulders MJ, Schijlen E, (2017) Re-sequencing transgenic plants revealed rearrangements at T-DNA inserts, and integration of a short T-DNA fragment, but no increase of small mutations elsewhere. Plant Cell Rep 36:493–504. https://doi.org/10.1007/s00299-017-2098-z
Seitz C, Vitten M, Steinbach P, Hartl S, Hirsche J, Rathje W, Forkmann G (2007) Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochemistry 68:824–833. https://doi.org/10.1016/j.phytochem.2006.12.012
Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Lo C (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228:1043–1054. https://doi.org/10.1007/s00425-008-0806-1
Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140:637–646. https://doi.org/10.1104/pp.105.072579
Streisfeld MA, Rausher MD (2011) Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution 65:629–642. https://doi.org/10.1111/j.1558-5646.2010.01165.x
Team RC (2015) R Foundation for Statistical Computing; Vienna, Austria: 2014. R: a language and environment for statistical computing
Wang GP, Yu XD, Sun YW, Jones HD, Xia LQ (2016) Generation of marker-and/or backbone-free transgenic wheat plants via Agrobacterium-mediated transformation. Front Plant Sci 7:1324. https://doi.org/10.3389/fpls.2016.01324
Weiss D (2000) Regulation of flower pigmentation and growth: multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiol Plantarum 110:152–157. https://doi.org/10.1034/j.1399-3054.2000.110202.x
Winkel-Shirley B (2001) It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol 127:1399–1404. https://doi.org/10.1104/pp.010675
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223. https://doi.org/10.1016/S1369-5266(02)00256-X
Winkel BS (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55:85–107. https://doi.org/10.1146/annurev.arplant.55.031903.141714
Yao SL, Xu Y, Zhang YY, Lu YH (2013) Black rice and anthocyanins induce inhibition of cholesterol absorption in vitro. Food Funct 4:1602–1608. https://doi.org/10.1039/c3fo60196j
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. J Plant Biotechnol 12:797–807. https://doi.org/10.1111/pbi.12200
Zhao C, Giusti MM, Malik M, Moyer MP, Magnuson BA (2004) Effects of commercial anthocyanin-rich extracts on colonic cancer and nontumorigenic colonic cell growth. J Agric Food Chem 52:6122–6128. https://doi.org/10.1021/jf049517a
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acid Res 42:10903–10914. https://doi.org/10.1093/nar/gku806
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01319302)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jung, Y.J., Lee, H.J., Kim, J.H. et al. CRISPR/Cas9-targeted mutagenesis of F3′H, DFR and LDOX, genes related to anthocyanin biosynthesis in black rice (Oryza sativa L.). Plant Biotechnol Rep 13, 521–531 (2019). https://doi.org/10.1007/s11816-019-00579-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-019-00579-4