Abstract
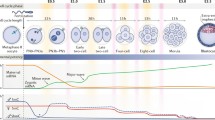
Extensive and dynamic chromatin remodeling occurs after fertilization, including DNA methylation and histone modifications. These changes underlie the transition from gametic to embryonic chromatin and are thought to facilitate early embryonic development. Histone H3 lysine 4 methylation (H3K4me) is an important epigenetic mechanism that associates with gene-specific activation and functions in development. However, dynamic regulation of H3K4me during early embryonic development remains unclear. Herein, the authors examined the dynamic changes of H3K4me and its key regulators (Ash1l, Ash2l, Kmt2a, Kmt2b, Kmt2c, Setd1a, Setd7, Kdm1a, Kdm1b, Kdm5a, Kdm5b, Kdm5c, and Kdm5d) in mouse oocytes and preimplantation embryos. An increase in levels of H3K4me2 and me3 was observed at the one- to two-cell stages (P < 0.05), corresponding to the period of embryonic genome activation (EGA). Subsequently, the H3K4me2 level dramatically decreased at the four-cell stage and remained at low level until the blastocyst stage (P < 0.05), whereas the H3K4me3 level transiently decreased in the four-cell embryos but steadily increased to the peak in the blastocysts (P < 0.05). The high level of H3K4me2 during the EGA was coinciding with a peak expression of its methyltransferase, ASH2L, which may stabilize this methylation level during this period. Correspondingly, a concomitant decrease in levels of its demethylases, KDM5B and KDM1A, was observed. H3K4me3 was correlated to the expression of its methyltransferase (KMT2B) and demethylase (KDM5A). Thus, these enzymes may function for the EGA and the first lineage segregation in preimplantation mouse embryos.




Similar content being viewed by others
References
Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K (2013) The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 9:e1003461
Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, Matzuk MM (2010) MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol 8:e1000453
Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145:183–197
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Beisel C, Paro R (2011) Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 12:123–135
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326
Black JC, Van Rechem C, Whetstine JR (2012) Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48:491–507
Chan SW, Hong W (2001) Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J Biol Chem 276:28402–28412
Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461:415–418
Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N (2007) Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol 17:808–812
Dillon SC, Zhang X, Trievel RC, Cheng X (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol 6:227
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13:713–719
Eissenberg JC, Shilatifard A (2010) Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 339:240–249
Eliazer S, Shalaby NA, Buszczak M (2011) Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc Natl Acad Sci U S A 108:7064–7069
Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG (2010) Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell 39:222–233
Gao Y, Hyttel P, Hall VJ (2010) Regulation of H3K27me3 and H3K4me3 during early porcine embryonic development. Mol Reprod Dev 77:540–549
Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A (2010) Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466:383–387
Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA (2007) Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol 27:8466–8479
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318
Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, Van Esch H, Kleefstra T, Hamel B, Moraine C, Gecz J, Turner G, Reinhardt R, Kalscheuer VM, Ropers HH, Lenzner S (2005) Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 76:227–236
Katz DJ, Edwards TM, Reinke V, Kelly WG (2009) A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137:308–320
Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7:715–727
Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, Lee SK, Chan L, Roeder RG, Lee JW (2008) Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci U S A 105:19229–19234
Lee JH, Skalnik DG (2008) Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28:609–618
Mamo S, Gal AB, Bodo S, Dinnyes A (2007) Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol 7:14
McGraw S, Vigneault C, Sirard MA (2007) Temporal expression of factors involved in chromatin remodeling and in gene regulation during early bovine in vitro embryo development. Reproduction 133:597–608
Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560
Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10:1107–1117
Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1:299–312
Pradhan S, Chin HG, Esteve PO, Jacobsen SE (2009) SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics 4:383–387
Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME (2010) Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 12:853–862
Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411
Sarmento OF, Digilio LC, Wang Y, Perlin J, Herr JC, Allis CD, Coonrod SA (2004) Dynamic alterations of specific histone modifications during early murine development. J Cell Sci 117:4449–4459
Shao GB, Ding HM, Gong AH (2008a) Role of histone methylation in zygotic genome activation in the preimplantation mouse embryo. In Vitro Cell Dev Biol Anim 44:115–120
Shao GB, Ding HM, Gong AH, Xiao DS (2008b) Inheritance of histone H3 methylation in reprogramming of somatic nuclei following nuclear transfer. J Reprod Dev 54:233–238
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953
Smith JA, White EA, Sowa ME, Powell ML, Ottinger M, Harper JW, Howley PM (2010) Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc Natl Acad Sci U S A 107:3752–3757
Stoller JZ, Huang L, Tan CC, Huang F, Zhou DD, Yang J, Gelb BD, Epstein JA (2010) Ash2l interacts with Tbx1 and is required during early embryogenesis. Exp Biol Med (Maywood) 235:569–576
Terranova R, Agherbi H, Boned A, Meresse S, Djabali M (2006) Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A 103:6629–6634
van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P (2005) Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev 122:1008–1022
VerMilyea MD, O’Neill LP, Turner BM (2009) Transcription-independent heritability of induced histone modifications in the mouse preimplantation embryo. PLoS One 4:e6086
Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W, He Q, Yang D, Chen R, Liu D, Barton M, Songyang Z (2013) The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem 288:5039–5048
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T (2009a) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41:125–129
Wang J, Zhang M, Zhang Y, Kou Z, Han Z, Chen DY, Sun QY, Gao S (2010) The histone demethylase JMJD2C is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol Reprod 82:105–111
Wang K, Chen Y, Chang EA, Knott JG, Cibelli JB (2009b) Dynamic epigenetic regulation of the Oct4 and Nanog regulatory regions during neural differentiation in rhesus nuclear transfer embryonic stem cells. Cloning Stem Cells 11:483–496
Wang SS, Zhou BO, Zhou JQ (2011) Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol Cell Biol 31:3171–3181
Whitworth KM, Agca C, Kim JG, Patel RV, Springer GK, Bivens NJ, Forrester LJ, Mathialagan N, Green JA, Prather RS (2005) Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol Reprod 72:1437–1451
Wu FR, Liu Y, Shang MB, Yang XX, Ding B, Gao JG, Wang R, Li WY (2012) Differences in H3K4 trimethylation in in vivo and in vitro fertilization mouse preimplantation embryos. Genet Mol Res 11:1099–1108
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD (2007) JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A 104:19226–19231
Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25:801–812
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81170573) and the Open Research Fund Program of Jiangsu Provincial Key Laboratory for Science and Technology of Photon Manufacturing (Jiangsu University) (GZ200710) and the Natural Science Foundation of Jiangsu Province (BK2011485).
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: T. Okamoto
Rights and permissions
About this article
Cite this article
Shao, GB., Chen, JC., Zhang, LP. et al. Dynamic patterns of histone H3 lysine 4 methyltransferases and demethylases during mouse preimplantation development. In Vitro Cell.Dev.Biol.-Animal 50, 603–613 (2014). https://doi.org/10.1007/s11626-014-9741-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-014-9741-6