Abstract
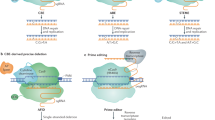
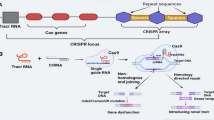
CRISPR-Cas technology has raised considerable interest among plant scientists, both in basic science and in plant breeding. Presently, the generation of random mutations at a predetermined site of the genome is well mastered, just like the targeted insertion of transgenes, although both remain restricted to species or genotypes amenable for plant transformation. On the other hand, true genome editing, i.e. the deliberate replacement of one or several nucleotides of the genome in a predetermined fashion, is limited to some rather particular examples that generally concern genes allowing positive selection, for example tolerance to herbicides. Therefore, further technological developments are necessary to fully exploit the potential of genome editing in enlarging the gene pool beyond the natural variability available in a given species. In principle, the technology can be applied to any quality related, agronomical or ecological trait, under the condition of upstream knowledge on the genes to be targeted and the precise modifications necessary to improve alleles. Published proof of concepts concern a wide range of agronomical traits, the most frequent being disease resistance, herbicide tolerance and the biochemical composition of harvested products. The regulatory status of the plants obtained by CRISPR-Cas technology raises numerous questions, in particular with regard to the plants that carry in their genomes the punctual modifications caused by the presence of the Cas9 nuclease but not the nuclease itself. Without clarification by the competent authorities, CRISPR-Cas technology would continue to be a powerful tool in functional genomics, but its potential in plant breeding would remain untapped.



Similar content being viewed by others
References
Ahloowalia BS, Maluszynski M, Nichterlein K (2004) Global impact of mutation-derived varieties. Euphytica 135:187–204
Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, Lemaux PG, Medford JI, Orozco-Cardenas ML, Tricoli DM, Van Eck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart CN Jr. (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28:1510–1520
Andersen MM, Landes X, Xiang W, Anyshchenko A, Falhof J, Osterberg JT, Olsen LI, Edenbrandt AK, Vedel SE, Thorsen BJ, Sandoe P, Gamborg C, Kappel K, Palmgren MG (2015) Feasibility of new breeding techniques for organic farming. Trends Plant Sci 20:426–434
Andersson M, Turesson H, Nicolia A, Falt AS, Samuelsson M, Hofvander P (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 36:117–128
Butler NM, Atkins PA, Voytas DF, Douches DS (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10:e0144591
Butler NM, Baltes NJ, Voytas DF, Douches DS (2016) Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front Plant Sci 7:1045
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17:1140–1153
Endo M, Mikami M, Toki S (2016) Biallelic gene targeting in rice. Plant Physiol 170:667–677
Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, Mathis L, Voytas DF, Zhang F (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J 12:934–940
Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 16:258
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco SC, Register IJ, Zhao ZY, Xu D, Jones TJ, Gordon-Kamm WJ (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28:1998–2015
Osterberg JT, Xiang W, Olsen LI, Edenbrandt AK, Vedel SE, Christiansen A, Landes X, Andersen MM, Pagh P, Sandoe P, Nielsen J, Christensen SB, Thorsen BJ, Kappel K, Gamborg C, Palmgren M (2017) Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci 22:373–384
Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J 15:1509–1519
Podevin N, Davies HV, Hartung F, Nogue F, Casacuberta JM (2013) Site-directed nucleases: a paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol 31:375–383
Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36:1–8
Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171:470–480 e478
Saito S, Maeda R, Adachi N (2017) Dual loss of human POLQ and LIG4 abolishes random integration. Nat Commun 8:16112
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15:207–216
Sprink T, Eriksson D, Schiemann J, Hartung F (2016) Regulatory hurdles for genome editing: process- vs. product-based approaches in different regulatory contexts. Plant Cell Rep 35:1493–1506
Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7:13274
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17103
Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X (2015) Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep 34:1473–1476
Wang X, Tu M, Wang D, Liu J, Li Y, Li Z, Wang Y, Wang X (2017) CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol J 16:844–855
Wang YP, Cheng X, Shan QW, Zhang Y, Liu JX, Gao CX, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Yin K, Gao C, Qiu JL (2017) Progress and prospects in plant genome editing. Nat Plants 3:17107
Zhou X, Zha M, Huang J, Li L, Imran M, Zhang C (2017) StMYB44 negatively regulates phosphate transport by suppressing expression of PHOSPHATE1 in potato. J Exp Bot 68:1265–1281
Funding
This study is funded by the Investissement d’Avenir program of the French National Agency of Research for the project GENIUS (ANR-11-BTBR-0001_GENIUS).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rogowsky, P.M. CRISPR-Cas Technology in Plant Science. Potato Res. 60, 353–360 (2017). https://doi.org/10.1007/s11540-018-9387-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11540-018-9387-y