Abstract
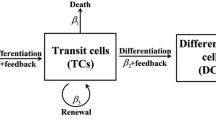
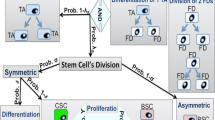
Mathematical modeling of cell differentiated in colonic crypts can contribute to a better understanding of basic mechanisms underlying colonic tissue organization, but also its deregulation during carcinogenesis and tumor progression. Here, we combined bifurcation analysis to assess the effect that time delay has in the complex interplay of stem cells and semi-differentiated cells at the niche of colonic crypts, and systematic model perturbation and simulation to find model-based phenotypes linked to cancer progression. The models suggest that stem cell and semi-differentiated cell population dynamics in colonic crypts can display chaotic behavior. In addition, we found that clinical profiling of colorectal cancer correlates with the in silico phenotypes proposed by the mathematical model. Further, potential therapeutic targets for chemotherapy resistant phenotypes are proposed, which in any case will require experimental validation.









Similar content being viewed by others
References
Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang T-W, Lasitschka F, Andrulis M et al (2013) A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 15:978–990
Adimy M, Crauste F, Ruan S (2006) Periodic oscillations in leukopoiesis models with two delays. J Theor Biol 242:288–299
Andronov AA, Chaikin CE (2012) Theory of oscillations. Ulan Press, Ulan
Arnold VI (1978) Ordinary differential equations. The MIT Press, Cambridge
Baker A-M, Cereser B, Melton S, Fletcher AG, Rodriguez-Justo M, Tadrous PJ, Humphries A, Elia G, McDonald SAC, Wright NA et al (2014) Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep 8:940–947
Barker N (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15:19–33
Berezkin M (1976) Comparison of the level of mitotic activity and duration of mitosis in normal and neoplastic mouse tissues during the 24-hour period. Bull Exp Biol Med 82(5):1704–1706
Broer H, Takens F (2010) Dynamical systems and chaos. Springer, Berlin
Carrato A (2008) Adjuvant treatment of colorectal cancer. Gastrointest Cancer Res GCR 2:S42–S46
Carulli AJ, Samuelson LC, Schnell S (2014) Unraveling intestinal stem cell behavior with models of crypt dynamics. Integr Biol Quant Biosci Nano Macro 6:243–257
Chicone C (2006) Ordinary differential equations with applications. Springer, Berlin
De Matteis G, Graudenzi A, Antoniotti M (2013) A review of spatial computational models for multi-cellular systems, with regard to intestinal crypts and colorectal cancer development. J Math Biol 66:1409–1462
Di Garbo A, Johnston M, Chapman S, Maini P (2010) Variable renewal rate and growth properties of cell populations in colon crypts. Phys Rev E 81:061909
Dingli D, Pacheco JM (2011) Stochastic dynamics and the evolution of mutations in stem cells. BMC Biol 9:41
Fletcher AG, Murray PJ, Maini PK (2015) Multiscale modelling of intestinal crypt organization and carcinogenesis. arXiv:1506.05019
Freedman HI, Rao VSH (1983) The trade-off between mutual interference and time lags in predator-prey systems. Bull Math Biol 45:991–1004
Granda T, Levi F (2002) Tumor-based rhythms of anticancer efficacy in experimental models. J Biol Med Rhythm Res 19(1):21–41
Hale J, Lunel S (1993) Introduction to functional differential equations. Springer, New York
Hayes N (1950) Roots of the transcendental equation associated with a certain differential difference equations. J Lond Math Soc 25:226–232
Hbid ML, Arino O, Dads EA (2008) Delay differential equations and applications: proceedings of the NATO advanced study institute held in Marrakech, Morocco, 9-21 September 2002 (Springer)
Hilborn R (2001) Chaos and nonlinear dynamics: an introduction for scientists and engineers. Oxford University Press, Oxford
Inagaki A, Sakata T (2005) Dose-dependent stimulatory and inhibitory effects of luminal and serosal n-butyric acid on epithelial cell proliferation of pig distal colonic mucosa. J Nutr Sci Vitaminol (Tokyo) 51:156–160
Johnston MD, Edwards CM, Bodmer WF, Maini PK, Chapman SJ (2007a) Mathematical modeling of cell population dynamics in the colonic crypt and in colorectal cancer. Proc Natl Acad Sci USA 104:4008–4013
Johnston MD, Edwards CM, Bodmer WF, Maini PK, Chapman SJ (2007b) Examples of mathematical modeling: tales from the crypt. Cell Cycle Georget Tex 6:2106–2112
Khan QJ, Greenhalgh D (1999) Hopf bifurcation in epidemic models with a time delay in vaccination. IMA J Math Appl Med Biol 16:113–142
Kilian H, Bartkowiak D, Kaufman D, Kemkemer R (2008) The general growth logistics of cell populations. Cell Biochem Biophys 51:51–66
Letellier C, Denis F, Aguirre LA (2013) What can be learned from a chaotic cancer model? J Theor Biol 322:7–16
Lombardo Y, Scopelliti A, Cammareri P, Todaro M, Iovino F, Ricci-Vitiani L, Gulotta G, Dieli F, de Maria R, Stassi G (2011) Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology 140:297–309
Lorentz O, Duluc I, Arcangelis AD, Simon-Assmann P, Kedinger M, Freund JN (1997) Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J Cell Biol 139:1553–1565
Mackey M, Glass L (1977) Oscillation and chaos in physiological control systems. Science New Ser 197:287–289
Marsden JE, McCracken M (1976) The Hopf bifurcation and its applications. Springer, New York
Monk NAM (2003) Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr Biol CB 13:1409–1413
Nikolov S (2013) Stability and Andronov–Hopf bifurcation of a system with three time delays. J Math 2013:e347071
Nikolov SG (2014) Modelling and analysis of miRNA regulation. BIOMATH 3:1407231
Nikolov S, Vera J, Kotev V, Wolkenhauer O, Petrov V (2008) Dynamic properties of a delayed protein cross talk model. Biosystems 91:51–68
Nikolov S, Ullah M, Nemov M, Vera J, Wolkenhauer O (2013) Modeling colorectal cancer: a stability analysis approach. In: Medical advancements in aging and regenerative technologies: clinical tools and applications (IGI Global)
Nikolov S, Wolkenhauer O, Vera J (2014) Tumors as chaotic attractors. Mol Biosyst 10:172–179
Nikolov S, Wolkenhauer O, Vera J (2015) Analysis of complex behavior of stem cell populations in colonic crypts. Int J Bioautom 19:S51–S68
Potten CS, Loeffler M (1990) Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Dev Camb Engl 110:1001–1020
Ritsma L, Ellenbroek SIJ, Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H, van Rheenen J (2014) Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507:362–365
Ruan S, Wei J (2003) On the zeros of transcendental functions with applications to stability of delay differential equations with two delays. Dyn Contin Discrete Impuls Syst Ser Math Anal 10:863–874
Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LCG, Lannon WA, Grotzinger C, Del Rio M et al (2013) A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 19:619–625
Sensse A, Eiswirth M (2005) Feedback loops for chaos in activator-inhibitor systems. J Chem Phys 122:44516
Shilnikov LP, Shilnikov AL, Turaev DV, Chua LO (1998) Methods of qualitative theory in nonlinear dynamics. World Scientific Pub Co Inc, Hackensack
Siegmund KD, Marjoram P, Tavaré S, Shibata D (2009) Many colorectal cancers are “flat” clonal expansions. Cell Cycle Georget Tex 8:2187–2193
Smallbone K, Corfe BM (2014) A mathematical model of the colon crypt capturing compositional dynamic interactions between cell types. Int J Exp Pathol 95:1–7
Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD et al (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143:134–144
Tomlinson IP, Bodmer WF (1995) Failure of programmed cell death and differentiation as causes of tumors: some simple mathematical models. Proc Natl Acad Sci USA 92:11130–11134
van Leeuwen IMM, Edwards CM, Ilyas M, Byrne HM (2007) Towards a multiscale model of colorectal cancer. World J Gastroenterol 13:1399–1407
Vera J, Schmitz U, Lai X, Engelmann D, Khan FM, Wolkenhauer O, Pützer BM (2013) Kinetic modeling-based detection of genetic signatures that provide chemoresistance via the E2F1-p73/DNp73-miR-205 network. Cancer Res 73:3511–3524
Vermeulen L, Morrissey E, van der Heijden M, Nicholson AM, Sottoriva A, Buczacki S, Kemp R, Tavaré S, Winton DJ (2013) Defining stem cell dynamics in models of intestinal tumor initiation. Science 342:995–998
Vogelstein B, Kinzler KW (2002) The genetic basis of human cancer. McGraw-Hill Professional, New York
Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D (2009) Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 9:489–499
Wodarz D, Komarova N (2005) Computational biology of cancer: lecture notes and mathematical modeling. WSPC, New York
Wolpin BM, Mayer RJ (2008) Systemic treatment of colorectal cancer. Gastroenterology 134:1296–1310
Zhao R, Michor F (2013) Patterns of proliferative activity in the colonic crypt determine crypt stability and rates of somatic evolution. PLOS Comput Biol 9:e1003082
Acknowledgements
This work was supported by the German Federal Ministry of Education and Research (BMBF) as part of the project e:Bio SysMet [0316171 to JV]. Julio Vera is funded by the Erlangen University Hospital (ELAN funds, 14-07-22-1-Vera-González and direct Faculty support).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Author Contributions
The initial idea, the mathematical model and the qualitative and numerical bifurcation analysis were developed by S. Nikolov. Mathematical simulations and in silico phenotypic signatures were computed by G Santos and J Vera. All the authors drafted the paper.
Appendix
Appendix
1.1 Andronov–Hopf Bifurcation for Second Fixed Points \(\bar{{N}}_0 \ne 0 ,\bar{{N}}_1 \ne 0.\)
The case \(\tau _1 >0,\;\tau _2 =0.\)
According to Nikolov et al. (2013), we assume that the finite time delay \(\tau _1 \) is longer than\(\tau _2 \). Setting \(\tau _2 =0\) in (14), the characteristic equation becomes
where \(K_{13} =K_1 -\psi _0 ,\quad K_{22} =K_2 -T_2 \) and \(T_{13} =T_1 -T_3 \).
It is well known that that the stability of the equilibrium state \(\bar{E}^{1}\) depends on the sign of the real parts of the roots of (A.1). We recall that a steady state is locally asymptotically stable if and only if all roots of (A.1) have negative real parts, and its stability can only be lost if these roots cross the vertical axis, that is if purely imaginary roots appear. Generally speaking, the transcendental equation (A.1) (for nonzero delay) cannot be solved analytically and has an indefinite number of roots. In essence, we have two main tools besides direct numerical integration: firstly, the linear stability analysis in the case of a small time delay, and secondly, the Hopf bifurcation theorem. From biological point of view it is known that time delay, \(\tau _1 \), is bigger than one (Nikolov et al. 2013) here we use Hopf bifurcation theorem. Thus, we let\(\chi =m+in \quad \left( {m,\;n\in R} \right) \) and rewrite (A.1) in terms of its real and imaginary parts as
To find the first bifurcation point, we look for purely imaginary roots\(\chi =\pm in\), \(n\in R\) of (A.1), i.e., we set \(m=0.\) Then the above two equations are reduced to
or another one
It is clear that if the first bifurcation point is \(\left( {n_b^0 ,\;\tau _b^0 } \right) \), then the other bifurcation points \(\left( {n_b ,\;\tau _b } \right) \) satisfy \(n_b \tau _b =n_b^0 \tau _b^0 +2\nu \pi , \quad \nu =1,\;2,\;\ldots ,\;\infty \).
One can notice that if n is a solution of (A.3) (or (A.4)), then so is \(-n\). Hence, in the following we only investigate for positive solutions n of (A.3) or (A.4), respectively. By squaring the two equations into system (A.2) and then adding them, it follows that:
As \(\bar{{E}^{1}}\) is locally asymptotically stable at \(\tau _1 =0\), it satisfies the Routh–Hurwitz conditions for stability for a square polynomial. Equation (A.5) is a square in \(n^{2}\), and the left-hand side is positive for very large values of \(n^{2}\) and negative for \(n=0\) if and only if \(T_{13}^2 >K_{22}^2 \), i.e., when Eq. (A.5) has at least one positive real root. Moreover, to apply the Hopf bifurcation theorem, according to Khan and Greenhalgh (1999), the following theorem in this situation applies:
Theorem 1
Suppose that \(n_b\) is the least positive simple root of (A.5). Then, \(in\left( {\tau _b } \right) =in_b\) is a simple root of (A.1) and \(m\left( {\tau _1 } \right) +in\left( {\tau _1 } \right) \) is differentiable with respect to \(\tau _1\) in a neighborhood of\(\tau _1 =\tau _b\).
To establish an Andronov–Hopf bifurcation at\(\tau _1 =\tau _b \), we need to show that a pair of complex eigenvalues crosses the imaginary axis with nonzero speed, i.e., the following transversally condition \(\left. {\frac{\hbox {d}\left( {Re\chi } \right) }{\hbox {d}t}} \right| _{\tau =\tau _b } \ne 0\) is satisfied.
From (A.3) we know that \(\tau _{b_k } \)corresponding to \(n_b\) is
Because for \(\tau _1 =0\), equilibrium \(\bar{E}^{1}\) is stable, by Butler’s lemma (Freedman and Rao 1983), equilibrium remains stable for \(\tau _1 <\tau _{b_k } \), where \(\tau _b =\tau _{b_k } \) as \(k=0.\) We have now to show that \(\left. {\frac{\hbox {d}\left( {Re\chi } \right) }{\hbox {d}t}} \right| _{\tau =\tau _b } \ne 0\).
Hence, if denote
then
Evaluating the real part of this equation at \(\tau _1 =\tau _b \) and setting \(\chi =in_b \) yield
where
Let \(\theta =n_b^2 \), then, (A.5) reduces to
Then, for \({g}'\left( \theta \right) \) we have
If \(n_b \) is the least positive simple root of (A.5), then
Hence,
According to the Hopf bifurcation theorem (Marsden and McCracken 1976), we define Theorem 2:
Theorem 2
If \(n_b \) is the least positive root of (A.5), then an Andronov–Hopf bifurcation occurs as \(\tau _1 \) passes through\(\tau _b \).
The case \(\tau _1>0,\;\tau _2 >0.\)
We return to the study of (14) with \(\tau _1 ,\;\tau _2 >0.\) In order to investigate the local stability of the equilibrium state \(\bar{{E}}^{1}\) of system (1), we first prove a result regarding the sign of the real parts of the characteristic roots of (14) in the next theorem:
Theorem 3
If all roots of (A.1) are with negative real parts for\(\tau _1 >0\) , then there exists a \(\tau _2^{bif} \left( {\tau _1 } \right) >0\) such that all roots of the characteristic equation (14) have negative real parts at\(\tau _2 <\tau _2^{bif} \left( {\tau _1 } \right) \) , i.e., when\(\tau _2 \in \left[ {0,\;\left. {\tau _2^{bif} \left( {\tau _1 } \right) } \right) } \right. \).
Here we omit the proof of Theorem 3 because it is similar to those in Adimy et al. (2006), Nikolov (2013).
Rights and permissions
About this article
Cite this article
Nikolov, S., Santos, G., Wolkenhauer, O. et al. Model-Based Phenotypic Signatures Governing the Dynamics of the Stem and Semi-differentiated Cell Populations in Dysplastic Colonic Crypts. Bull Math Biol 80, 360–384 (2018). https://doi.org/10.1007/s11538-017-0378-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-017-0378-y