Abstract
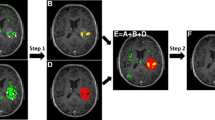
We address the problem of fully automated region discovery and robust image segmentation by devising a new deformable model based on the level set method (LSM) and the probabilistic nonnegative matrix factorization (NMF). We describe the use of NMF to calculate the number of distinct regions in the image and to derive the local distribution of the regions, which is incorporated into the energy functional of the LSM. The results demonstrate that our NMF–LSM method is superior to other approaches when applied to synthetic binary and gray-scale images and to clinical magnetic resonance images (MRI) of the human brain with and without a malignant brain tumor, glioblastoma multiforme. In particular, the NMF–LSM method is fully automated, highly accurate, less sensitive to the initial selection of the contour(s) or initial conditions, more robust to noise and model parameters, and able to detect as small distinct regions as desired. These advantages stem from the fact that the proposed method relies on histogram information instead of intensity values and does not introduce nuisance model parameters. These properties provide a general approach for automated robust region discovery and segmentation in heterogeneous images. Compared with the retrospective radiological diagnoses of two patients with non-enhancing grade 2 and 3 oligodendroglioma, the NMF–LSM detects earlier progression times and appears suitable for monitoring tumor response. The NMF–LSM method fills an important need of automated segmentation of clinical MRI.
















Similar content being viewed by others
References
Babalola KO, Patenaude B, Aljabar P, Schnabel J, Kennedy D, Crum W, Smith S, Cootes TF, Jenkinson M, Rueckert D (2008) Comparison and evaluation of segmentation techniques for subcortical structures in brain MRI. Med Image Comput Comput Assist Interv 5241:409–416
Bayar B, Bouaynaya N, Shterenberg R (2014) Probabilistic non-negative matrix factorization: theory and application to microarray data analysis. J Bioinform Comput Biol 12:25
Caselles V, Kimmel R, Sapiro G (1997) Geodesic active contours. Int J Comput Vis 22(1):61–79
Chan T, Vese L (2001) Active contours without edges. IEEE Trans Image Process 10:266–277
Chen Y, Zhang J, Mishra A, Yang J (2011) Image segmentation and bias correction via an improved level set method. Neurocomputing 74(17):3520–3530
Chena Y, Zhanga J, Macioneb J (2009) An improved level set method for brain mr images segmentation and bias correction. Comput Med Imaging Graph 33:510–519
Chuang KS, Tzeng HL, Chen S, Wu J, Chen TJ (2006) Fuzzy c-means clustering with spatial information for image segmentation. Comput Med Imaging Graph 30:9–15
Cohen LD, Cohen I (1993) Finite-element methods for active contour models and balloons for 2-D and 3-D images. IEEE Trans Pattern Anal Mach Intell 15(11):1131–1147
Friston KJ, Ashburner J, Frith C, Poline JB, Heather JD, Frackowiak RS (1995) Spatial registration and normalization of images. Hum Brain Mapp 2:165–189
Garcia-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL (2013) Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 17(1):1–18
Jain S, Sima DM, Ribbens A, Cambron M, Maertens A, Van Hecke W, De Mey J, Barkhof F, Steenwijk MD, Daams M, Maes F, Van Huffel S, Vrenken H, Smeets D (2015) Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. Neuroimage Clin 8:367–375
Kass M, Witkin A, Terzopoulos D (1987) Snakes: active contour models. Int J Comput Vis 1:321–331
Klauschen F, Goldman A, Barra V, Meyer-Lindenberg A, Lundervold A (2009) Evaluation of automated brain MR image segmentation and volumetry methods. Hum Brain Mapp 30(4):1310–1327
Li C, Kao CY, Gore JC, Ding Z (2008) Minimization of region-scalable fitting energy for image segmentation. IEEE Trans Image Process 17(10):1940–1949
Li C, Xu C, Gui C, Fox MD (2010) Distance regularized level set evolution and its application to image segmentation. IEEE Trans Image Process 19:3242–3254
Li C, Huang R, Ding Z, Gatenby JC, Metaxas DN, Gore JC (2011) A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI. IEEE Trans Image Process 20(7):2007–2016
Pagnozzi AM, Gal Y, Boyd RN, Fiori S, Fripp J, Rose S, Dowson N (2015) The need for improved brain lesion segmentation techniques for children with cerebral palsy: a review. Int J Dev Neurosci 47:229–246
Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-berg H, Bannister PR, Luca MD, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, Stefano ND, Brady JM, Matthews PM (2004) Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23:208–219
Tsai A, Yezzi A, Willsky AS (2001) Curve evolution implementation of the Mumford–Shah functional for image segmentation, denoising, interpolation and magnification. IEEE Trans Image Process 10:1169–1186
Valverde S, Oliver A, Diez Y, Cabezas M, Vilanova JC, Ramio-Torrenta L, Rovira A, Llado X (2015) Evaluating the effects of white matter multiple sclerosis lesions on the volume estimation of 6 brain tissue segmentation methods. AJNR Am J Neuroradiol 36(6):1109–1115
Vovk U, Pernus F, Likar B (2007) A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging 26:405–421
Wang L, He L, Mishra A, Li C (2009) Active contours driven by local Gaussian distribution fitting energy. Signal Process 89(12):2435–2447
Acknowledgments
This work is supported by the National Science Foundation under Award Number ACI-1429467. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect those of the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dera, D., Bouaynaya, N. & Fathallah-Shaykh, H.M. Automated Robust Image Segmentation: Level Set Method Using Nonnegative Matrix Factorization with Application to Brain MRI. Bull Math Biol 78, 1450–1476 (2016). https://doi.org/10.1007/s11538-016-0190-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-016-0190-0