Abstract
A model of mutation rate evolution for multiple loci under arbitrary selection is analyzed. Results are obtained using techniques from Karlin (Evolutionary Biology, vol. 14, pp. 61–204, 1982) that overcome the weak selection constraints needed for tractability in prior studies of multilocus event models.
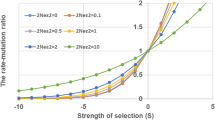
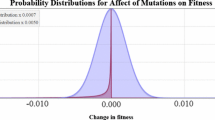
A multivariate form of the reduction principle is found: reduction results at individual loci combine topologically to produce a surface of mutation rate alterations that are neutral for a new modifier allele. New mutation rates survive if and only if they fall below this surface—a generalization of the hyperplane found by Zhivotovsky et al. (Proc. Natl. Acad. Sci. USA 91, 1079–1083, 1994) for a multilocus recombination modifier. Increases in mutation rates at some loci may evolve if compensated for by decreases at other loci. The strength of selection on the modifier scales in proportion to the number of germline cell divisions, and increases with the number of loci affected. Loci that do not make a difference to marginal fitnesses at equilibrium are not subject to the reduction principle, and under fine tuning of mutation rates would be expected to have higher mutation rates than loci in mutation-selection balance.
Other results include the nonexistence of ‘viability analogous, Hardy–Weinberg’ modifier polymorphisms under multiplicative mutation, and the sufficiency of average transmission rates to encapsulate the effect of modifier polymorphisms on the transmission of loci under selection. A conjecture is offered regarding situations, like recombination in the presence of mutation, that exhibit departures from the reduction principle. Constraints for tractability are: tight linkage of all loci, initial fixation at the modifier locus, and mutation distributions comprising transition probabilities of reversible Markov chains.
Similar content being viewed by others
References
Ababneh, F., Jermiin, L. S., & Robinson, J. (2006). Generation of the exact distribution and simulation of matched nucleotide sequences on a phylogenetic tree. J. Math. Model. Algorithms, 5, 291–308.
Altenberg, L. (1984). A generalization of theory on the evolution of modifier genes. Ph.D. thesis, Stanford University. Searchable online and available from University Microfilms, Ann Arbor, MI.
Altenberg, L. (2009). The evolutionary reduction principle for linear variation in genetic transmission. Bull. Math. Biol., 71, 1264–1284.
Altenberg, L., & Feldman, M. W. (1987). Selection, generalized transmission, and the evolution of modifier genes. I. The reduction principle. Genetics, 117, 559–572.
Baer, C. F., Miyamoto, M. M., & Denver, D. R. (2007). Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet., 8, 619–631.
Balkau, B., & Feldman, M. W. (1973). Selection for migration modification. Genetics, 74, 171–174.
Brandon, R. N. (1982). The levels of selection. In P. Asquith & T. Nickles (Eds.), PSA 1982 (Vol. 1, pp. 315–323). East Lansing: Philosophy of Science Association.
Charlesworth, B. (1990). Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res., 55, 199–221.
Charlesworth, B., & Charlesworth, D. (1979). Selection on recombination in clines. Genetics, 91, 581–589.
Charlesworth, B., Charlesworth, D., & Strobeck, C. (1979). Selection for recombination in partially self-fertilizing populations. Genetics, 93, 237–244.
Deutsch, E., & Neumann, M. (1984). Derivatives of the Perron root at an essentially nonnegative matrix and the group inverse of an M-matrix. J. Math. Anal. Appl., 102, 1–29.
Duistermaat, J. J., & Kolk, J. A. C. (2004). Cambridge studies in advanced mathematics: Vol. 86. Multidimensional real analysis I: Differentiation. Cambridge: Cambridge University Press. ISBN 9780521551144.
Eyre-Walker, A., & Keightley, P. D. (2007). The distribution of fitness effects of new mutations. Nat. Rev. Genet., 8, 610–618.
Feldman, M. W. (1972). Selection for linkage modification: I. Random mating populations. Theor. Popul. Biol., 3, 324–346.
Feldman, M. W., & Balkau, B. (1973). Selection for linkage modification II. A recombination balance for neutral modifiers. Genetics, 74, 713–726.
Feldman, M. W., & Krakauer, J. (1976). Genetic modification and modifier polymorphisms. In S. Karlin & E. Nevo (Eds.), Population genetics and ecology (pp. 547–583). New York: Academic Press.
Feldman, M. W., & Liberman, U. (1986). An evolutionary reduction principle for genetic modifiers. Proc. Natl. Acad. Sci. USA, 83, 4824–4827.
Feldman, M. W., Christiansen, F. B., & Brooks, L. D. (1980). Evolution of recombination in a constant environment. Proc. Natl. Acad. Sci. USA, 77, 4838–4841.
Feller, W. (1971). An introduction to probability theory and its applications, Vol. I (3rd ed.). New York: Wiley.
Fox, A., Tuch, B., & Chuang, J. (2008). Measuring the prevalence of regional mutation rates: an analysis of silent substitutions in mammals, fungi, and insects. BMC Evol. Biol., 8, 186.
Giraud, A., Matic, I., Tenaillon, O., Clara, A., Radman, M., Fons, M., & Taddei, F. (2001). Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science, 291.
Guillemin, V., & Pollack, A. (1974). Differential topology. Prentice-Hall: Englewood Cliffs.
Hirsch, M. W. (1976). Differential topology. New York: Springer.
Hoede, C., Denamur, E., & Tenaillon, O. (2006). Selection acts on DNA secondary structures to decrease transcriptional mutagenesis. PLoS Genet., 2, e176. http://dx.plos.org/10.1371%2Fjournal.pgen.0020176.
Holsinger, K., Feldman, M. W., & Altenberg, L. (1986). Selection for increased mutation rates with fertility differences between matings. Genetics, 112, 909–922.
Holsinger, K. E., & Feldman, M. W. (1983a). Linkage modification with mixed random mating and selfing: a numerical study. Genetics, 103, 323–333.
Holsinger, K. E., & Feldman, M. W. (1983b). Modifiers of mutation rate: evolutionary optimum with complete selfing. Proc. Natl. Acad. Sci. USA, 80, 6732–6734.
Horn, R. A., & Johnson, C. R. (1985). Matrix analysis. Cambridge: Cambridge University Press.
Iosifescu, M. (1980). Finite Markov processes and their applications. Bucharest: Wiley.
Jayaswal, V., Jermiin, L. S., & Robinson, J. (2005). Estimation of phylogeny using a general Markov model. Evol. Bioinform. Online, 1, 62–80.
Karlin, S. (1976). Population subdivision and selection migration interaction. In S. Karlin & E. Nevo (Eds.), Population genetics and ecology (pp. 616–657). New York: Academic Press.
Karlin, S. (1982). Classification of selection-migration structures and conditions for a protected polymorphism. In M. K. Hecht, B. Wallace, & G. T. Prance (Eds.), Evolutionary biology (Vol. 14, pp. 61–204). New York: Plenum.
Karlin, S., & McGregor, J. (1972a). Application of method of small parameters to multi-niche population genetic models. Theor. Popul. Biol., 3, 186–209.
Karlin, S., & McGregor, J. (1972b). The evolutionary development of modifier genes. Proc. Natl. Acad. Sci. USA, 69, 3611–3614.
Karlin, S., & McGregor, J. (1974). Towards a theory of the evolution of modifier genes. Theor. Popul. Biol., 5, 59–103.
Keilson, J. (1979). Markov chain models: rarity and exponentiality. New York: Springer.
King, D. G., & Kashi, Y. (2007). Mutation rate variation in eukaryotes: evolutionary implications of site-specific mechanisms. Nat. Rev. Genet., 8.
Kingman, J. F. C. (1978). A simple model for the balance between selection and mutation. J. Appl. Probab., 15, 1–12.
Kingman, J. F. C. (1980). Mathematics of genetic diversity. Philadelphia: Society for Industrial and Applied Mathematics. ISBN 0-89871-166-5.
Kondrashov, A. S. (1982). Selection against harmful mutations in large sexual and asexual populations. Genet. Res., 40, 325–332.
Kondrashov, A. S. (1984). Deleterious mutations as an evolutionary factor. I. The advantage of recombination. Genet. Res., 44, 199–217.
Kondrashov, A. S. (1995). Modifiers of mutation-selection balance: general approach and the evolution of mutation rates. Genet. Res., 66, 53–69.
Kondrashov, F. A., & Kondrashov, A. S. (2010). Measurements of spontaneous rates of mutations in the recent past and the near future. Philos. Trans. R. Soc. B, 365, 1169–1176.
Lewontin, R. C. (1974). The genetic basis of evolutionary change. New York: Columbia University Press.
Liberman, U., & Feldman, M. W. (1986a). A general reduction principle for genetic modifiers of recombination. Theor. Popul. Biol., 30, 341–371.
Liberman, U., & Feldman, M. W. (1986b). Modifiers of mutation rate: A general reduction principle. Theor. Popul. Biol., 30, 125–142.
Lynch, M. (2010). Rate molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA, 107, 961–968.
Lynch, M., Sung, W., Morris, K., Coffey, N., Landry, C. R., Dopman, E. B., Dickinson, W. J., Okamoto, K., Kulkarni, S., Hartl, D. L., & Thomas, W. K. (2008). A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci., 105, 9272–9277.
Munkres, J. R. (1975). Topology: a first course. Prentice-Hall: Englewood Cliffs. ISBN 0-13-925495-1.
Otto, S. P., & Feldman, M. W. (1997). Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol., 51, 34–47.
Pylkov, K. V., Zhivotovsky, L. A., & Feldman, M. W. (1998). Migration versus mutation in the evolution of recombination under multilocus selection. Genet. Res., 71, 247–256.
Roach, J. C., Glusman, G., Smit, A. F. A., Huff, C. D., Hubley, R., Shannon, P. T., Rowen, L., Pant, K. P., Goodman, N., Bamshad, M., Shendure, J., Drmanac, R., Jorde, L. B., Hood, L., & Galas, D. J. (2010). Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. http://dx.doi.org/10.1126/science.1186802.
Rodríguez, F., Oliver, J., Marín, A., & Medina, J. (1990). The general stochastic model of nucleotide substitution. J. Theor. Biol., 142, 485–501.
Salmon, W. C. (1971). Statistical explanation and statistical relevance. Pittsburgh: University of Pittsburgh Press.
Salmon, W. C. (1984). Scientific explanation and the causal structure of the world. Princeton: Princeton University Press.
Singer, I. M., & Thorpe, J. A. (1967). Lecture notes on elementary topology and geometry. New York: Springer. ISBN 0-387-90202-3.
Squartini, F., & Arndt, P. F. (2008). Quantifying the stationarity and time reversibility of the nucleotide substitution process. Mol. Biol. Evol., 25, 2525–2535.
Teague, R. (1977). A model of migration modification. Theor. Popul. Biol., 12, 86–94.
Whelan, S., & Goldman, N. (2004). Estimating the frequency of events that cause multiple-nucleotide changes. Genetics, 167, 2027–2043.
Wilkinson, J. H. (1965). The algebraic eigenvalue problem. Oxford: Clarendon Press.
Yang, Z. (1995). On the general reversible Markov process model of nucleotide substitution: a reply to Saccone et al. J. Mol. Evol., 41, 254–255.
Yang, Z., & Nielsen, R. (2002). Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol., 19, 908–917.
Zhivotovsky, L. A., & Feldman, M. W. (1995). The reduction principle for recombination under density-dependent selection. Theor. Popul. Biol., 47, 244–256.
Zhivotovsky, L. A., Feldman, M. W., & Christiansen, F. B. (1994). Evolution of recombination among multiple selected loci: A generalized reduction principle. Proc. Natl. Acad. Sci. USA, 91, 1079–1083.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to the memory of Sam Karlin, whose theorems continue to bear new fruit.
Rights and permissions
About this article
Cite this article
Altenberg, L. An Evolutionary Reduction Principle for Mutation Rates at Multiple Loci. Bull Math Biol 73, 1227–1270 (2011). https://doi.org/10.1007/s11538-010-9557-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-010-9557-9