Abstract
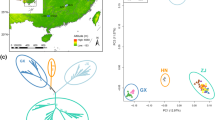
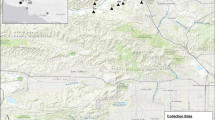
Genome-wide DArTseq scans of 268 individuals of Eucalyptus salubris, distributed along an aridity gradient in southwestern Australia, revealed cryptic population structure that appears to signal hitherto unappreciated ecotypic differentiation and barriers to gene flow. Genome-wide scans were undertaken on 30 wild-sampled individuals from each of nine populations; 10 individuals per population were measured for habit and functional traits. DArTseq generated 16,122 high-quality markers, of which 56.3 % located to E. grandis chromosomes. Genetic affinities of the nine populations were only weakly correlated with geographic distances. Rather, populations appeared to form two distinct molecular lineages that maintained their distinctiveness in an area of geographic overlap. Twenty-four outlier markers signalled divergent selection and differentiation of the two putative lineages. Populations from the two lineages were phenotypically differentiated in leaf thickness, specific leaf area (SLA) and leaf nitrogen per unit mass (Nmass). The more northerly lineage (with thinner leaves) occurred in hotter, drier conditions with higher radiation. Populations of the more southerly lineage occurred on soils that were relatively low in phosphorus; the trees had thicker leaves, lower SLA and lower leaf Nmass, consistent with general responses to low nutrient levels. While historic isolation and drift may have contributed to the cryptic population structure observed, there is evidence of ecotypic adaptation, which may provide an exogenous barrier to gene flow. This study highlights the power of new molecular technologies to provide novel insights into the genetic architecture of wild populations.




Similar content being viewed by others
References
Abdelaziz M, Lorite J, Munoz-Pajares AJ, Herrador MB, Perfectti F, Gomez JM (2011) Using complementary techniques to distinguish cryptic species: a new Erysimum (Brassicaceae) species from North Africa. Am J Bot 98:1049–1060. doi:10.3732/ajb.1000438
Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F (2004) Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. doi:10.1093/bioinformatics/btg427
Anderson MJ, Gorley RN, Clarke KR (2008) Permanova + for primer: guide to software and statistical methods. PRIMER-E, Plymouth
Australian Plant Name Index (2014) Eucalyptus salubris F. Muell. Centre for Plant Biodiversity Research, Australian Government
Barbour RC et al (2009) A geographic mosaic of genetic variation within a foundation tree species and its community-level consequences. Ecology 90:1762–1772. doi:10.1890/08-0951.1
Barrett CF, Freudenstein JV (2011) An integrative approach to delimiting species in a rare but widespread mycoheterotrophic orchid. Mol Ecol 20:2771–2786. doi:10.1111/j.1365-294X.2011.05124.x
Bickford D et al (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155. doi:10.1016/j.tree.2006.11.004
Bierne N, Welch J, Loire E, Bonhomme F, David P (2011) The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol 20:2044–2072. doi:10.1111/j.1365-294X.2011.05080.x
Bomblies K (2010) Doomed lovers: mechanisms of isolation and incompatibility in plants. Annu Rev Plant Biol 61:109–124. doi:10.1146/annurev-arplant-042809-112146
Brooker MIH (2000) A new classification of the genus Eucalyptus L’Her. (Myrtaceae). Aust Syst Bot 13:79–148. doi:10.1071/sb98008
Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A (2012) Inferring species trees directly from Biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol 29:1917–1932. doi:10.1093/molbev/mss086
Byrne M (2008) Phylogeny, diversity and evolution of eucalypts. In: Ch. 11 in Sharma AK and Sharma A (eds) Plant genome biodiversity and evolution. Part E. phanerogams - angiosperms. Science Publishers, Berlin. pp 303–346
Byrne M, Moran GF, Tibbits WN (1993) Restriction map and maternal inheritance of chloroplast DNA in Eucalyptus nitens. J Hered 84:218–220
Carstens BC, Pelletier TA, Reid NM, Satler JD (2013) How to fail at species delimitation. Mol Ecol 22:4369–4383. doi:10.1111/mec.12413
Castro-Díez P, Villar-Salvador P, Pérez-Rontomé C, Maestro-Martínez M, Montserrat-Martí G (1997) Leaf morphology and leaf chemical composition in three Quercus (Fagaceae) species along a rainfall gradient in NE Spain. Trees Struct Funct 11:127–134. doi:10.1007/pl00009662
Clarke KR, Gorley RN (2006) Primer v6: user manual/tutorial. PRIMER-E, Plymouth
Crespo A, Perez-Ortega S (2009) Cryptic species and species pairs in lichens: a discussion on the relationship between molecular phylogenies and morphological characters. An Jard Bot Madr 66:71–81. doi:10.3989/ajbm.2225
de Queiroz LP, Lavin M (2011) Coursetia (Leguminosae) from eastern Brazil: nuclear ribosomal and chloroplast DNA sequence analysis reveal the monophyly of three caatinga-inhabiting species. Syst Bot 36:69–79. doi:10.1600/036364411x553144
de Stefano RD, Fernandez-Concha GC, Can-Itza LL, Lavin M (2010) The morphological and phylogenetic distinctions of Coursetia greenmanii (Leguminosae): taxonomic and ecological implications. Syst Bot 35:289–295
Derieg NJ, Sangaumphai A, Bruederle LP (2008) Genetic diversity and endemism in North American Carex section Ceratocystis (Cyperaceae). Am J Bot 95:1287–1296. doi:10.3732/ajb.2007069
Derieg NJ, Weil SJ, Reznicek AA, Bruederle LP (2013) Carex viridistellata sp. nov. (Cyperaceae), a new cryptic species from prairie fens of the eastern United States. Syst Bot 38:82–91. doi:10.1600/036364413x661971
Desper R, Gascuel O (2002) Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol 9:687–705. doi:10.1089/106652702761034136
Dettman JR, Jacobson DJ, Taylor JW (2003) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703–2720. doi:10.1554/03-073
Dyer RJ, Nason JD (2004) Population graphs: the graph theoretic shape of genetic structure. Mol Ecol 13:1713–1727. doi:10.1111/j.1365-294X.2004.02177.x
Esau K (1960) Anatomy of seed plants. Wiley, New York
Felsenstein J (2006) Accuracy of coalescent likelihood estimates: do we need more sites, more sequences, or more loci? Mol Biol Evol 23:691–700
Fine PVA, Garcia-Villacorta R, Pitman NCA, Mesones I, Kembel SW (2010) A floristic study of the white-sand forests of Peru. Ann Mo Bot Gard 97:283–305. doi:10.3417/2008068
Foll M, Gaggiotti O (2008) A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
French M (ed) (2012) Eucalypts of Western Australia’s wheatbelt. Malcolm French, Padbury
Fujita MK, Leache AD, Burbrink FT, McGuire JA, Moritz C (2012) Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol Evol 27:480–488. doi:10.1016/j.tree.2012.04.012
Goodstein DM et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186. doi:10.1093/nar/gkr944
Greve C, Gimnich F, Hutterer R, Misof B, Haase M (2012) Radiating on oceanic islands: patterns and processes of speciation in the land snail genus Theba (Risso 1826). PLoS ONE 7:e34339. doi:10.1371/journal.pone.0034339
Guerin GR, Wen HX, Lowe AJ (2012) Leaf morphology shift linked to climate change. Biol Lett 8:882–886. doi:10.1098/rsbl.2012.0458
Hedenas L et al (2014) Three species for the price of one within the moss Homalothecium sericeum s.l. Taxon 63:249–257
Hill RS (1998) Fossil evidence for the onset of xeromorphy and scleromorphy in Australian Proteaceae. Aust Syst Bot 11:391–400. doi:10.1071/sb97016
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi:10.1093/bioinformatics/17.8.754
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalo grass, Buchloe dactyloides (Nutt.) Engelm. Theor Appl Genet 86:927–934. doi:10.1007/bf00211043
Huson D, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Johnson LAS, Hill KD (1991) Systematic studies in the eucalypts 2. A revision of the gimlets and related species: Eucalyptus extracodical series Salubres and Annulatae (Myrtaceae). Telopea 4:201–222
Jordan GJ, Hill RS (1996) The fossil record of the Epacridaceae. Ann Bot 77:341–346. doi:10.1006/anbo.1996.0041
Knowles LL, Carstens BC (2007) Estimating a geographically explicit model of population divergence. Evolution 61:477–493. doi:10.1111/j.1558-5646.2007.00043.x
Ladiges P, Parra-O C, Gibbs A, Udovicic F, Nelson G, Bayly M (2011) Historical biogeographical patterns in continental Australia: congruence among areas of endemism of two major clades of eucalypts. Cladistics 27:29–41. doi:10.1111/j.1096-0031.2010.00315.x
Lamont BB, Groom PK, Cowling RM (2002) High leaf mass per area of related species assemblages may reflect low rainfall and carbon isotope discrimination rather than low phosphorus and nitrogen concentrations. Funct Ecol 16:403–412. doi:10.1046/j.1365-2435.2002.00631.x
Leavitt SD, Fankhauser JD, Leavitt DH, Porter LD, Johnson LA, St Clair LL (2011) Complex patterns of speciation in cosmopolitan “rock posy” lichens—discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol Phylogenet Evol 59:587–602. doi:10.1016/j.ympev.2011.03.020
Leliaert F, Verbruggen H, Vanormelingen P, Steen F, Lopez-Bautista JM, Zuccarello GC, De Clerck O (2014) DNA-based species delimitation in algae. Eur J Phycol 49:179–196. doi:10.1080/09670262.2014.904524
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi:10.1093/nar/gkr201
Liu L, Pearl DK (2007) Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst Biol 56:504–514. doi:10.1088/10635150701429982
Lousada JM, Lovato MB, Borba EL (2013) High genetic divergence and low genetic variability in disjunct populations of the endemic Vellozia compacta (Velloziaceae) occurring in two edaphic environments of Brazilian campos rupestres. Braz J Bot 36:45–53. doi:10.1007/s40415-013-0001-x
Macfarlane C, Warren CR, White DA, Adams MA (1999) A rapid and simple method for processing wood to crude cellulose for analysis of stable carbon isotopes in tree rings. Tree Physiol 19:831–835
McKinnon GE, Vaillancourt RE, Steane DA, Potts BM (2004) The rare silver gum, Eucalyptus cordata, is leaving its trace in the organellar gene pool of Eucalyptus globulus. Mol Ecol 13:3751–3762. doi:10.1111/j.1365-294X.2004.02364.x
McLean EH, Prober SM, Stock WD, Steane DA, Potts BM, Vaillancourt RE, Byrne M (2014) Plasticity of functional traits varies clinally along a rainfall gradient in Eucalyptus tricarpa. Plant Cell Environ 37:1440–1451. doi:10.1111/pce.12251
Medina R, Lara F, Goffinet B, Garilleti R, Mazimpaka V (2012) Integrative taxonomy successfully resolves the pseudo-cryptic complex of the disjunct epiphytic moss Orthotrichum consimile s.l. (Orthotrichaceae). Taxon 61:1180–1198
Menz MHM, Phillips RD, Anthony JM, Bohman B, Dixon KW, Peakall R (2015) Ecological and genetic evidence for cryptic ecotypes in a rare sexually deceptive orchid, Drakaea elastica. Bot J Linn Soc 177:124–140. doi:10.1111/boj.12230
Mills PJ, Cook LG (2014) Rapid chromosomal evolution in a morphologically cryptic evolution. Mol Phylogenet Evol 77:126–135
Molina MD, Divakar PK, Millanes AM, Sanchez E, Del-Prado R, Hawksworth DL, Crespo A (2011) Parmelia sulcata (Ascomycota: Parmeliaceae), a sympatric monophyletic species complex. Lichenologist 43:585–601. doi:10.1017/s0024282911000521
Myburg AA et al (2014) The genome of Eucalyptus grandis. Nature 510:356–362. doi:10.1038/nature13308
Nicolle D (2006) A classification and census of regenerative strategies in the eucalypts (Angophora, Corymbia and Eucalyptus-Myrtaceae), with special reference to the obligate seeders. Aust J Bot 54:391–407. doi:10.1071/bt05061
Otalora MAG, Martinez I, Aragon G, Molina MC (2010) Phylogeography and divergence date estimates of a lichen species complex with a disjunct distribution pattern. Am J Bot 97:216–223. doi:10.3732/ajb.0900064
Pandey KK (1977) Mentor pollen: possible role of wall-held pollen growth promoting substances in overcoming intra- and interspecific incompatibility. Genetica 47:219–229
Pannell JR, Fields PD (2014) Evolution in subdivided plant populations: concepts, recent advances and future directions. New Phytol 201:417–432. doi:10.1111/nph.12495
Peakall R, Smouse P (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pennington RT, Daza A, Reynel C, Lavin M (2011) Poissonia eriantha (Leguminosae) From Cuzco, Peru: an overlooked species underscores a pattern of narrow endemism common to seasonally dry neotropical vegetation. Syst Bot 36:59–68. doi:10.1600/036364411x553135
Pillon Y, Hopkins HCF, Munzinger J, Amir H, Chase MW (2009) Cryptic species, gene recombination and hybridization in the genus Spiraeanthemum (Cunoniaceae) from New Caledonia. Bot J Linn Soc 161:137–152
Pizano C, Mangan SA, Herre EA, Eom AH, Dalling JW (2011) Above- and belowground interactions drive habitat segregation between two cryptic species of tropical trees. Ecology 92:47–56. doi:10.1890/09-1715.1
Porter SS (2013) Adaptive divergence in seed color camouflage in contrasting soil environments. New Phytol 197:1311–1320. doi:10.1111/nph.12110
Read J, Sanson GD, de Garine-Wichatitsky M, Jaffre T (2006) Sclerophylly in two contrasting tropical environments: low nutrients vs. low rainfall. Am J Bot 93:1601–1614. doi:10.3732/ajb.93.11.1601
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi:10.1093/bioinformatics/btg180
Sansaloni CP, Petroli CD, Jaccoud D, Carling J, Detering F, Grattapaglia D, Kilian A (2011) Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc 5:P54
Sarkinen TE, Marcelo-Pena JL, Yomona AD, Simon MF, Pennington RT, Hughes CE (2011) Underestimated endemic species diversity in the dry inter-Andean valley of the Rio Maranon, northern Peru: an example from Mimosa (Leguminosae, Mimosoideae). Taxon 60:139–150
Schimper AFW (1903) Plant geography upon a physiological basis (English translation by W.R. Fisher). Clarendon, Oxford
Schulze ED, Turner NC, Nicolle D, Schumacher J (2006) Leaf and wood carbon isotope ratios, specific leaf areas and wood growth of Eucalyptus species across a rainfall gradient in Australia. Tree Physiol 26:479–492
Seehausen O et al (2014) Genomics and the origin of species. Nat Rev Genet 15:176–192. doi:10.1038/nrg3644
Sexton JP, Hangartner SB, Hoffmann AA (2014) Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68:1–15. doi:10.1111/evo.12258
Silva JCE, Potts B, Tilyard P (2012) Epistasis causes outbreeding depression in eucalypt hybrids. Tree Genet Genomes 8:249–265. doi:10.1007/s11295-011-0437-8
Skabo S, Vaillancourt RE, Potts BM (1998) Fine-scale genetic structure of Eucalyptus globulus ssp. globulus forest revealed by RAPDs. Aust J Bot 46:583–594. doi:10.1071/bt97056
Slomka A, Sutkowska A, Szczepaniak M, Malec P, Mitka J, Kuta E (2011) Increased genetic diversity of Viola tricolor L. (Violaceae) in metal-polluted environments. Chemosphere 83:435–442. doi:10.1016/j.chemosphere.2010.12.081
Sotuyo S, Delgado-Salinas A, Chase MW, Lewis GP, Oyama K (2007) Cryptic speciation in the Caesalpinia hintonii complex (Leguminosae: Caesalpinioideae) in a seasonally dry Mexican forest. Ann Bot 100:1307–1314. doi:10.1093/aob/mcm213
Sotuyo S, Lewis GP (2007) A new species of Caesalpinia from the Rio Balsas Depression, Mexico, and an updated taxonomic circumscription of the Caesalpinia hintonii complex (Leguminosae: Caesalpinioideae: Caesalpinieae: Poincianella Group). Brittonia 59:33–36. doi:10.1663/0007-196x(2007)59[33:ansocf]2.0.co;2
Steane DA, Nicolle D, McKinnon GE, Vaillancourt RE, Potts BM (2002) Higher-level relationships among the eucalypts are resolved by ITS-sequence data. Aust Syst Bot 15:49–62. doi:10.1071/sb00039
Steane DA, Potts BM, McLean E, Prober SM, Stock WD, Vaillancourt RE, Byrne M (2014) Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol Ecol 23:2500–2513. doi:10.1111/mec.12751
Strasburg JL, Sherman NA, Wright KM, Moyle LC, Willis JH, Rieseberg LH (2012) What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos Trans R Soc B Biol Sci 367:364–373. doi:10.1098/rstb.2011.0199
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th edn. Sinauer Associates, Sunderland
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. doi:10.1006/fgbi.2000.1228
Tellier F, Tapia J, Faugeron S, Destombe C, Valero M (2011) The Lessonia nigrescent species complex (Laminariales, Phaeophyceae) shows strict parapatry and complete reproductive isolation in a secondary contact zone. J Phycol 47:894–903. doi:10.1111/j.1529-8817.2011.01019.x
Verhoeven KJF, Simonsen KL, McIntyre LM (2005) Implementing false discovery rate control: increasing your power. Oikos 108:643–647. doi:10.1111/j.0030-1299.2005.13727.x
Warren CR, Adams MA (2005) What determines interspecific variation in relative growth rate of Eucalyptus seedlings? Oecologia 144:373–381. doi:10.1007/s00442-005-0092-6
Warren CR, Dreyer E, Tausz M, Adams MA (2006) Ecotype adaptation and acclimation of leaf traits to rainfall in 29 species of 16-year-old Eucalyptus at two common gardens. Funct Ecol 20:929–940. doi:10.1111/j.1365-2435.2006.01198.x
Warren CR, Tausz M, Adams MA (2005) Does rainfall explain variation in leaf morphology and physiology among populations of red ironbark (Eucalyptus sideroxylon subsp tricarpa) grown in a common garden? Tree Physiol 25:1369–1378
Wolf MA, Sciuto K, Andreoli C, Moro I (2012) Ulva (Chlorophyta, Ulvales) biodiversity in the North Adriatic Sea (Mediterranean, Italy): cryptic species and new introductions. J Phycol 48:1510–1521. doi:10.1111/jpy.12005
Woodhams M, Steane DA, Jones RC, Nicolle D, Moulton V, Holland BR (2013) Novel distances for Dollo data. Syst Biol 62:62–77. doi:10.1093/sysbio/sys071
Wright IJ, Westoby M (2002) Leaves at low versus high rainfall: coordination of structure, lifespan and physiology. New Phytol 155:403–416. doi:10.1046/j.1469-8137.2002.00479.x
Xu T, Hutchinson M (2011) ANUCLIM version 6.1 user guide. The Australian National University, Fenner School of Environment and Society, Canberra
Acknowledgments
This work was funded by a grant from the Australian National Climate Change Adaptation Research Facility (TB11 03) with additional support through the Great Western Woodlands Supersite of Australia’s Terrestrial Ecosystem Research Network and ARC Discovery Grant DP130104220. We thank: Craig Macfarlane, Nat Raisbeck-Brown, Georg Wiehl, Didier Alanoix (CSIRO Land and Water Flagship) and Anne Rick for assistance with field work; Bronwyn MacDonald, Rachel Binks and Donna Bradbury (Department of Parks and Wildlife, Western Australia), Carl Gosper (CSIRO Land and Water Flagship and Department of Parks and Wildlife, Western Australia), Paul Tilyard and Chris Burridge (University of Tasmania) for technical assistance; the project’s end-user advisory group, David Freudenberger, Gary Howling, Neil Riches and Richard Mazanec; the Western Australian Herbarium for accession of samples and assistance with and access to FloraBase data; and the Tasmanian Partnership for Advanced Computing (TPAC).
Data archiving statement
Data relating to this study is available from Dryad: doi:10.5061/dryad.h06r3. These include Climate data, soil data, isotope data (relating to physiology), morphometric data and dominant (presence/absence) DArTseq data (including DNA sequences of each DArTseq marker).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Grattapaglia
Topical Collection on Population structure
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resources 1
(DOCX 23 kb)
Online Resources 2
(DOCX 50 kb)
Online Resources 3
(DOCX 16 kb)
Online Resources 4
(PDF 995 kb)
Rights and permissions
About this article
Cite this article
Steane, D.A., Potts, B.M., McLean, E. et al. Genome-wide scans reveal cryptic population structure in a dry-adapted eucalypt. Tree Genetics & Genomes 11, 33 (2015). https://doi.org/10.1007/s11295-015-0864-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0864-z