Abstract
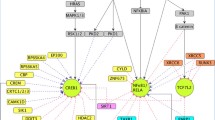
We present an approach to extracting information from textual documents of biological knowledge and demonstrate how cellular gene pathways may be inferred. Natural language processing techniques are used to represent title and abstract fields of publications to derive a gene similarity vectors which are subject to cluster analysis. Gene interactions are derived by parsing sentences in the abstracts to infer causal relationships. We show how high throughput transcriptome data may then be used to enhance the construction of gene pathways from information derived from text. Subnetworks constructed by integrating information automatically derived from literature with gene expression data is validated by comparing biological processes defined in the Gene Ontology 2(GO) database. We find that precision increases in \(58\%\) of the clusters when enhanced in this manner while a decrease in precision is observed in a relatively small number of clusters. These results are compared to similar attempts at the same problem and appear to be better in terms of precision of network construction. We also show an example of a subnetwork found by this analysis that overlaps a known gene pathway in KEGG and MIPS databases.






Similar content being viewed by others
References
Benthem, J. F., & Meulen, A. G. (1997). Handbook of logic and language. Elsevier.
Corney, D. P. A., Buxton, B. F., Langdon, W. B., & Jones, D. T. (2004). BioRAT: Extracting biological information from full-length papers. Bioinformatics, 20(17), 3206–3213.
Brown, M. S. P., Grundy, W. N., Lin, D., Cristianini, N., Sugnet, C. W., Furey, T. S., et al. (2000). Knowledge-based analysis of microarray gene expression data by using support vector machines. Proceedings of the National Academy of Sciences of the USA, 97(1), 262–267.
Eisen, M. B., Spellman, P. T., Brown, P. O., & Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the USA, 95, 14863–14868.
Grossman, D., & Frieder, O. (1999). Introduction to modern information retrieval. London: Library Association Publishing.
Iliopoulos, I., Enright, A., & Ouzounis, C. (2001). Textquest: Document clustering of medline abstracts for concept discovery in molecular biology. Pac. Symp. Biocomput., 199, 384–395.
Jenssen, T., Laegreid, A., Komorowski, J., & Hovig, R. (2001). A literature network of human genes for high-throughput analysis of gene expression. Nature Genetics, 28, 21–28.
Kanehisa, M., Goto, S., Kawashima, S., & Nakaya, A. (2002). The KEGG databases at GenomeNet. Nucleic Acids Research, 30(1), 42–46.
Karopka, T., Scheel, T., Bansemer, S., & Glass, A. (2004). Automatic construction of gene relation networks using text mining and gene expression data. Medical Informatics and the Internet in Medicine, 29(2), 169-183.
Mering, C. V., Zdobnov, E. M., Tsoka, S., Ciccarelli, F. D., Pereira-Leal, J. B., Ouzounis, C. A., et al. (2003). Genome evolution reveals biochemical networks and functional modules. Proceedings of the National Academy of Sciences of the USA, 100(26), 15428–15433.
Pavlidis, P., & Grundy, W. N. (2000). Combining Microarray Expression Data and Phylogenetic Profiles to Learn Gene Functional Categories Using Support Vector Machines. Technical report, Columbia University Department of Computer Science.
Raychaudhuri, S., Schutze, H., & Altman, R. B. (2003). Inclusion of textual documentation in the analysis of multidimensional data sets: Application to gene expression data. Machine Learning, 52, 119–145.
Salton, G., & Buckley, C. (1988). Term weighting approaches in automatic text retrieval. Information Processing and Management, 24(5), 513–523.
Schlitt, T., Palin, K., Rung, J., Dietmann, S., Lappe, M., Ukkonen, E., et al. (2003). From gene networks to gene function. Genome Research, 13, 2568–2576.
Schultz, J. M., & Liberman, M. (1999). Topic detection and tracking using idf-weighted cosine coefficient. Proceedings of the DARPA Broadcast News Workshop, pp. 189–192.
Schwikowski, B., Uetz, P., & Fields, S. (2000). A network of protein–protein interactions in yeast. Nature Biotechnology, 18, 1257–1261.
Sekimizu, T., Park, H., & Tsujii, J. (1998). Identifying the interaction between genes and gene products based on frequently seen verbs in medline abstracts. Genome Informatics, 9, 62–71.
Stein, L. (2003). Integrating biological databases. Nature, 4, 337–345.
Tamayo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., et. al. (1999). Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proceedings of the National Academy of Sciences of the USA, 96, 2907–2912.
Acknowledgement
We are grateful to the Language Technology Group, the University of Edinburgh (LT CHUNK) and AT&T Labs-Research (Graphviz), SGD (GoTermFinder) for making software available in the public domain. SS was funded by The Royal Thai Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suwannaroj, S., Niranjan, M. Enhancing Automatic Construction of Gene Subnetworks by Integrating Multiple Sources of Information. J Sign Process Syst Sign Image 50, 331–340 (2008). https://doi.org/10.1007/s11265-007-0148-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11265-007-0148-4