Abstract
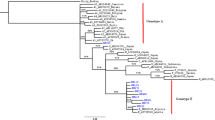
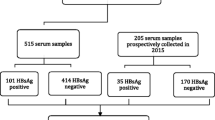
Hepatitis B virus (HBV) poses a significant threat to blood transfusion safety in sub-Saharan Africa (SSA) where allogeneic blood donations are screened serologically, and more sensitive nucleic acid tests (NATs) are utilized infrequently. HBV strains circulating among blood donors in Botswana are not yet characterized. We designed a cross-sectional study to determine the HBV sub-genotypes and prevalence of hepatitis B surface antigen (HBsAg) among blood donors between November 2014 and October 2015. A total of 12,575 blood donations were screened for HBsAg and 50 consecutive plasma samples were selected for genotyping from confirmed HBsAg+ donations. Overlapping Pol and complete S (Pol/S) open reading frames (ORFs) were sequenced from extracted HBV DNA. To identify any signature amino acids, mutations were compared to sequences from a cohort of chronic HBV patients co-infected with HIV and were treatment naïve. The prevalence of HBsAg+ blood donors was 1.02% (95% CI 0.9–1.2%), and the circulating sub-genotypes were A1 serotype adw2 (36.1%), D2 serotype ayw2 (2.9%), and D3 serotypes ayw 1/2 (58.3%). Prevalence of escape mutations was 14% from HBV isolates of blood donors and 15% from isolates of HBV/HIV co-infected patients (p = 0.6926). The escape mutations sP120L, sG130R, sY134H, and sD144A were identified predominantly among HBV isolates from blood donors. These escape mutations have been associated with accelerated HBV sequelae [e.g., liver cirrhosis (LC) and hepatocellular carcinoma (HCC)], failure to detect HBsAg, inability to respond to immunoglobulin (Ig) therapy, and HBV vaccine escape. Characterizing the HBV burden, circulating sub-genotypes, and clinically relevant mutations among blood donors in Botswana is important to elucidate the efficacy of currently available vaccines, predicting HBV-transmission patterns, understanding the cohort’s risk to HBV-related complications, and to developing prevention strategies and effective genotype-based antiretroviral therapies.




Similar content being viewed by others
References
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386(10003):1546–1555. https://doi.org/10.1016/S0140-6736(15)61412-X
Spearman CW, Afihene M, Ally R, Apica B, Awuku Y, Cunha L, Dusheiko G, Gogela N, Kassianides C, Kew M, Lam P, Lesi O, Lohoues-Kouacou MJ, Mbaye PS, Musabeyezu E, Musau B, Ojo O, Rwegasha J, Scholz B, Shewaye AB, Tzeuton C, Sonderup MW, Gastroenterology, Hepatology Association of sub-Saharan A (2017) Hepatitis B in sub-Saharan Africa: strategies to achieve the 2030 elimination targets. Lancet Gastroenterol Hepatol 2(12):900–909. https://doi.org/10.1016/S2468-1253(17)30295-9
Franco E, Bagnato B, Marino MG, Meleleo C, Serino L, Zaratti L (2012) Hepatitis B: epidemiology and prevention in developing countries. World J Hepatol 4(3):74–80. https://doi.org/10.4254/wjh.v4.i3.74
Beguelin C, Fall F, Seydi M, Wandeler G (2018) The current situation and challenges of screening for and treating hepatitis B in sub-Saharan Africa. Expert Rev Gastroenterol Hepatol 12(6):537–546. https://doi.org/10.1080/17474124.2018.1474097
Jayaraman S, Chalabi Z, Perel P, Guerriero C, Roberts I (2010) The risk of transfusion-transmitted infections in sub-Saharan Africa. Transfusion 50(2):433–442. https://doi.org/10.1111/j.1537-2995.2009.002402.x
WHO (2012) Blood donor selection: guidelines on assessing donor suitability for blood donation. World Health Organization, Geneva.
WHO (2017) Blood safety and donation World Health Organisation http://www.who.int/mediacentre/factsheets/fs279/en/index.html. Accessed 04 March 2018
Vermeulen M, Swanevelder R, Chowdhury D, Ingram C, Reddy R, Bloch EM, Custer BS, Murphy EL (2017) Use of blood donor screening to monitor prevalence of HIV and hepatitis B and C viruses, South Africa. Emerg Infect Dis 23(9):1560–1563. https://doi.org/10.3201/eid2309.161594
WHO (2017) Current status on blood safety and availability in the WHO African Region—report of the 2013 survey. WHO Regional Office for Africa, Brazzaville
Apata IW, Averhoff F, Pitman J, Bjork A, Yu J, Amin NA, Dhingra N, Kolwaite A, Marfin A, Centers for Disease C, Prevention (2014) Progress toward prevention of transfusion-transmitted hepatitis B and hepatitis C infection–sub-Saharan Africa, 2000–2011. MMWR Morb Mortal Wkly Rep 63(29):613–619
Sunbul M (2014) Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol 20(18):5427–5434. https://doi.org/10.3748/wjg.v20.i18.5427
Coppola N, Onorato L, Minichini C, Di Caprio G, Starace M, Sagnelli C, Sagnelli E (2015) Clinical significance of hepatitis B surface antigen mutants. World J Hepatol 7(27):2729–2739. https://doi.org/10.4254/wjh.v7.i27.2729
Locarnini SA (1998) Hepatitis B virus surface antigen and polymerase gene variants: potential virological and clinical significance. Hepatology 27(1):294–297. https://doi.org/10.1002/hep.510270144
Kramvis A (2014) Genotypes and genetic variability of hepatitis B virus. Intervirology 57(3–4):141–150. https://doi.org/10.1159/000360947
Anderson M, Gaseitsiwe S, Moyo S, Wessels MJ, Mohammed T, Sebunya TK, Powell EA, Makhema J, Blackard JT, Marlink R, Essex M, Musonda RM (2015) Molecular characterisation of hepatitis B virus in HIV-1 subtype C infected patients in Botswana. BMC Infect Dis 15:335. https://doi.org/10.1186/s12879-015-1096-4
Matthews PC, Beloukas A, Malik A, Carlson JM, Jooste P, Ogwu A, Shapiro R, Riddell L, Chen F, Luzzi G, Jaggernath M, Jesuthasan G, Jeffery K, Ndung’u T, Goulder PJ, Geretti AM, Klenerman P (2015) Prevalence and characteristics of hepatitis B virus (HBV) coinfection among HIV-positive women in South Africa and Botswana. PLoS ONE 10(7):e0134037. https://doi.org/10.1371/journal.pone.0134037
Mbangiwa T, Kasvosve I, Anderson M, Thami PK, Choga WT, Needleman A, Phinius BB, Moyo S, Leteane M, Leidner J, Blackard JT, Mayondi G, Kammerer B, Musonda RM, Essex M, Lockman S, Gaseitsiwe S (2018) Chronic and occult hepatitis B virus infection in pregnant women in Botswana. Genes (Basel). https://doi.org/10.3390/genes9050259
Sequencher® DNA Sequence Analysis Software. http://www.genecodes.com. Accessed 10 Oct 2017
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29(8):1969–1973. https://doi.org/10.1093/molbev/mss075
Schultz AK, Bulla I, Abdou-Chekaraou M, Gordien E, Morgenstern B, Zoaulim F, Deny P, Stanke M (2012) jpHMM: recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res 40(Web Server issue):W193–W198. https://doi.org/10.1093/nar/gks414
Bell TG, Kramvis A (2015) Bioinformatics tools for small genomes, such as hepatitis B virus. Viruses 7(2):781–797. https://doi.org/10.3390/v7020781
Korber B, Myers G (1992) Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retrovir 8(9):1549–1560. https://doi.org/10.1089/aid.1992.8.1549
Chevalier MS, Kuehnert M, Basavaraju SV, Bjork A, Pitman JP (2016) Progress toward strengthening national blood transfusion services—14 Countries, 2011–2014. MMWR Morb Mortal Wkly Rep 65(5):115–119. https://doi.org/10.15585/mmwr.mm6505a4
Vermeulen M, Swanevelder R, Chowdhury D, Ingram C, Reddy R, Bloch EM, Custer BS, Murphy EL, Epidemiology NR, Donor evaluation Study IIIIC (2017) Use of blood donor screening to monitor prevalence of HIV and hepatitis B and C viruses, South Africa. Emerg Infect Dis 23(9):1560–1563. https://doi.org/10.3201/eid2309.161594
Mavenyengwa RT, Mukesi M, Chipare I, Shoombe E (2014) Prevalence of human immunodeficiency virus, syphilis, hepatitis B and C in blood donations in Namibia. BMC Public Health 14:424. https://doi.org/10.1186/1471-2458-14-424
Aydin OA, Karaosmanoglu HK, Sayan M, Ince ER, Nazlican O (2015) Seroprevalence and risk factors of syphilis among HIV/AIDS patients in Istanbul, Turkey. Cent Eur J Public Health 23(1):65–68. https://doi.org/10.21101/cejph.a4001
Kao JH, Chen PJ, Lai MY, Chen DS (2000) Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118(3):554–559
Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, Okita K, Okanoue T, Iino S, Tanaka E, Suzuki K, Watanabe H, Hige S, Mizokami M (2001) Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 34(3):590–594. https://doi.org/10.1053/jhep.2001.27221
Makondo E, Bell TG, Kramvis A (2012) Genotyping and molecular characterization of hepatitis B virus from human immunodeficiency virus-infected individuals in southern Africa. PLoS ONE 7(9):e46345. https://doi.org/10.1371/journal.pone.0046345
Hubschen JM, Andernach IE, Muller CP (2008) Hepatitis B virus genotype E variability in Africa. J Clin Virol 43(4):376–380. https://doi.org/10.1016/j.jcv.2008.08.018
Hou J, Liu Z, Gu F (2005) Epidemiology and prevention of hepatitis B virus infection. Int J Med Sci 2(1):50–57
Hannachi N, Bahri O, Ben Fredj N, Boukadida J, Triki H (2010) Risk of vertical transmission of hepatitis B virus in Tunisia. Arch Inst Pasteur Tunis 87(1–2):17–24
Baha W, Ennaji MM, Lazar F, Melloul M, El Fahime E, El Malki A, Bennani A (2012) HBV genotypes prevalence, precore and basal core mutants in Morocco. Infect Genet Evol 12(6):1157–1162. https://doi.org/10.1016/j.meegid.2012.04.026
Zhu HL, Li X, Li J, Zhang ZH (2016) Genetic variation of occult hepatitis B virus infection. World J Gastroenterol 22(13):3531–3546. https://doi.org/10.3748/wjg.v22.i13.3531
Amini-Bavil-Olyaee S, Vucur M, Luedde T, Trautwein C, Tacke F (2010) Differential impact of immune escape mutations G145R and P120T on the replication of lamivudine-resistant hepatitis B virus e antigen-positive and -negative strains. J Virol 84(2):1026–1033. https://doi.org/10.1128/JVI.01796-09
Lazarevic I (2014) Clinical implications of hepatitis B virus mutations: recent advances. World J Gastroenterol 20(24):7653–7664. https://doi.org/10.3748/wjg.v20.i24.7653
Coppola N, Loquercio G, Tonziello G, Azzaro R, Pisaturo M, Di Costanzo G, Starace M, Pasquale G, Cacciapuoti C, Petruzziello A (2013) HBV transmission from an occult carrier with five mutations in the major hydrophilic region of HBsAg to an immunosuppressed plasma recipient. J Clin Virol 58(1):315–317. https://doi.org/10.1016/j.jcv.2013.06.020
Salpini R, Colagrossi L, Bellocchi MC, Surdo M, Becker C, Alteri C, Aragri M, Ricciardi A, Armenia D, Pollicita M, Di Santo F, Carioti L, Louzoun Y, Mastroianni CM, Lichtner M, Paoloni M, Esposito M, D’Amore C, Marrone A, Marignani M, Sarrecchia C, Sarmati L, Andreoni M, Angelico M, Verheyen J, Perno CF, Svicher V (2015) Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immunosuppression. Hepatology 61(3):823–833. https://doi.org/10.1002/hep.27604
Zaaijer HL, Torres P, Ontanon A, Ponte LG, Koppelman MH, Lelie PN, Hemert FJ, Boot HJ (2008) Multiple surface antigen mutations in five blood donors with occult hepatitis B virus infection. J Med Virol 80(8):1344–1349. https://doi.org/10.1002/jmv.21233
Svicher V, Cento V, Bernassola M, Neumann-Fraune M, Van Hemert F, Chen M, Salpini R, Liu C, Longo R, Visca M, Romano S, Micheli V, Bertoli A, Gori C, Ceccherini-Silberstein F, Sarrecchia C, Andreoni M, Angelico M, Ursitti A, Spano A, Zhang JM, Verheyen J, Cappiello G, Perno CF (2012) Novel HBsAg markers tightly correlate with occult HBV infection and strongly affect HBsAg detection. Antiviral Res 93(1):86–93. https://doi.org/10.1016/j.antiviral.2011.10.022
Meldal BH, Bon AH, Prati D, Ayob Y, Allain JP (2011) Diversity of hepatitis B virus infecting Malaysian candidate blood donors is driven by viral and host factors. J Viral Hepat 18(2):91–101. https://doi.org/10.1111/j.1365-2893.2010.01282.x
El Chaar M, El Jisr T, Allain JP (2012) Hepatitis B virus DNA splicing in Lebanese blood donors and genotype A to E strains: implications for hepatitis B virus DNA quantification and infectivity. J Clin Microbiol 50(10):3159–3167. https://doi.org/10.1128/JCM.01251-12
Greer AE, Ou SS, Wilson E, Piwowar-Manning E, Forman MS, McCauley M, Gamble T, Ruangyuttikarn C, Hosseinipour MC, Kumarasamy N, Nyirenda M, Grinsztejn B, Pilotto JH, Kosashunhanan N, Goncalves de Melo M, Makhema J, Akelo V, Panchia R, Badal-Faesen S, Chen YQ, Cohen MS, Eshleman SH, Thio CL, Valsamakis A (2017) Comparison of hepatitis B virus infection in HIV-infected and HIV-uninfected participants enrolled in a multinational clinical trial: HPTN 052. J Acquir Immune Defic Syndr 76(4):388–393. https://doi.org/10.1097/QAI.0000000000001511
Garmiri P, Rezvan H, Abolghasemi H, Allain JP (2011) Full genome characterization of hepatitis B virus strains from blood donors in Iran. J Med Virol 83(6):948–952. https://doi.org/10.1002/jmv.21772
Acknowledgements
The authors would like to acknowledge the support and collaborations from National Blood Transfusions Services of Botswana. We also acknowledge the Ministry of Health and Wellness of Botswana for granting the permissions to conduct this pioneer study for molecular epidemiology of HBV in blood donors in Botswana. We would like to also extend our acknowledgements to the National University of Science and Technology, Zimbabwe; University of Botswana; and the Botswana-Harvard HIV Reference Laboratory for their sponsorship, support, and contribution to the success of the study.
Funding
This work was supported through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [Grant No. DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [Grant No. 107752/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust, or the UK government.
Author information
Authors and Affiliations
Contributions
WTC wrote the first draft of the manuscript. WTC and MA collaborated in the lab work, primary data analysis. SM and PM were involved in statistical analysis. TM and BBP were involved in conducting some of the lab work and manuscript editing before submission. EZ, TKS, and IK supervised the project and edited the manuscript prior to submission and data evaluation. MKK provided with samples and their demographics, and also edited the manuscript before submission. JTB developed the secondary analysis plan and performed the phylogenetics of the study. ME and RMM edited the manuscript prior to submission and complemented it with contextual data. SG developed the study, main directions of the analysis plan, overall interpretation of results and edited manuscript before submission. All co-authors read and authorized the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The participants’ samples used in the study were residual from blood donors, obtained already de-identified and anonymous. These were a sub-population of HBV + screened samples from National Blood Transfusion services in Botswana (NBTS). The study was approved by the University of Botswana Institute Review Board and the Health Research Development Division (HRDD) at the Botswana Ministry of Health and Wellness (HPDME 13/18/1).
Additional information
Edited by Wolfram Gerlich.
Rights and permissions
About this article
Cite this article
Choga, W.T., Anderson, M., Zumbika, E. et al. Molecular characterization of hepatitis B virus in blood donors in Botswana. Virus Genes 55, 33–42 (2019). https://doi.org/10.1007/s11262-018-1610-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-018-1610-z