Abstract
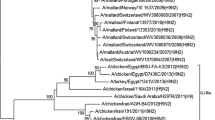
An outbreak of highly pathogenic avian influenza A (H5N1) virus in poultry was reported from Nandurbar and Jalgaon districts of Maharashtra and adjoining areas of Uchhal in Gujarat and Burhanpur in Madhya Pradesh in India from January to April, 2006. In the present study, the full genome of two previously uncharacterized strains of H5N1 viruses isolated at the National Institute of Virology (NIV), Pune, from post-mortem tissues of chicken collected from Navapur, Nandurbar district during the outbreak, has been presented. All the genes belong to clade 2.2 of the Z genotype and are close to the 2006 isolates from Iran, Afghanistan, Mongolia, Italy, and Krasnodar. In a study reported earlier, based on the partial gene sequences of HA, the authors (Pattnaik et al.) hypothesized that the viruses in Jalgaon and Navapur, causing outbreaks 12 days apart, were introduced at different times from different sources. However, our Navapur isolates are closer to the isolate reported from Jalgaon than that from Navapur. Molecular markers suggest that the isolates are sensitive to both drugs Oseltamivir and Amantadine. Amino acid residues responsible for pathogenesis, glycosylation, and receptor binding have also been discussed. The relationship between the Indian viruses and those in the East Africa/West-Asia flyway of migratory birds and the position of Nandurbar in this route suggests that the viruses in India may have been introduced through migratory birds although the role of trade as a possible route of introduction of the virus cannot be ruled out.


Similar content being viewed by others
References
X. Xu, K. Subbarao, N.J. Cox, Y. Guo, Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261, 15–19 (1999)
K.S. Li, Y. Guan, J. Wang et al., Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430, 209–213 (2004)
H. Chen, G.J.D. Smith, S.Y. Zhang, K. Qin, J. Wang, K.S. Li, et al., Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192 (2005)
World Organization for Animal Health (2006) Update on avian influenza in animals (Type H5), available at http://www.oie.int/downld/avianinfluenza/A_AI-Asia.htm. Accessed 21 Nov 2006
H. Chen, Y. Li, Z. Li, J. Shi, K. Shinya, G. Deng et al., Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in Western China. J. Virol. 80, 5976–5983 (2006)
G.J. Smith, X.H. Fan, J. Wang, et al., Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U.S.A. 103, 16936–16941 (2006)
Avian Influenza in India, Follow-up report No. 4 (final report) http://www.oie.int/eng/info/hebdo/AIS_05.HTM#Sec5. Vol. 19, No. 33, Accessed 17 Aug 2006
B. Pattnaik, A.K. Pateriya, R. Khandia, C. Tosh, S. Nagarajan, S. Gounalan, et al., Phylogenetic analysis revealed genetic similarity of the H5N1 avian influenza viruses isolated from HPAI outbreaks in chickens in Maharashtra, India with those isolated from swan in Italy and Iran in 2006. Curr. Sci. 91, 77–81 (2006)
R. Webster, N. Cox, K. Stöhr, Laboratory procedures, in WHO Manual on Animal Influenza Diagnosis and Surveillance, 2nd edn. 15–65 (2002)
A.P. Kendal, M.S. Pereira, J.J. Skehel, Hemagglutinin inhibition. in Concepts and Procedures for Laboratory-Based Influenza Surveillance, ed. by A.P. Kendal, M.S. Pereira, J.J. Skehel, (Centers for Disease Control and Prevention an Pan American Health Organization, Atlanta, 1982), pp. B17–B35
Recommended laboratory tests to identify avian influenza A virus in specimens from humans. WHO June 2005. Available at http://www.who.int/csr/disease/avian_influenza/guidelines/avian-labtests2.pdf. Accessed 21 Nov 2006
E. Hoffmann, J. Stech, Y. Guan, R.G. Webster, D.R. Perez, Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146, 2275–2289 (2001)
The World Health Organization Global Influenza Program Surveillance Network, Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 11, 1515–1521 (2005)
D.G. Higgins, J.D. Thompson, T.J. Gibson, Using CLUSTAL for multiple sequence alignments. Meth. Enzymol. 2666, 383–402 (1996)
S. Kumar, K. Tamura, M. Nei, MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (2004)
D.L. Swofford, PAUP*: Phylogenetic Analysis Using Parsimony (and other methods) 4.0 Beta. (Sinauer Associates, Sunderland, MA, 2002)
E. De Castro, C.J.A. Sigrist, A. Gattiker, V. Bulliard, S. Petra, Langendijk-Genevaux PS et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 (2006)
J. Stevens, O. Blixt, T.M. Tumpey, J.K. Taubenberger, J.C. Paulson, I.A. Wilson, Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–412 (2006)
R.J. Russell, L.F. Haire, D.J. Stevens, P.J. Collins, Y.P. Lin, G.M. Blackburn, A.J. Hay, S.J. Gamblin, J.J. Skehel, The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443, 45–49 (2006)
A. Sali, T.L. Blundell, Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
N. Guex M.C. Peitsch, SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714 (1997)
M. Shaw L. Cooper, X. Xu et al., Molecular changes associated with the transmission of avian influenza A H5N1 and H9N2 viruses to humans. J. Med. Virol. 66, 107–114 (2002)
G. Gabriel, B. Dauber, T. Wolff, O. Planz, H.D. Klenk, J. Stech, The viral polymerase mediates adaptation of an avian inßuenza virus to a mammalian host. Proc. Natl. Acad. Sci. U.S.A. 102, 18590–18595 (2005)
J.M. Katz, X. Lu, T.M. Tumpey, C.B. Smith, M.W. Shaw, K. Subbarao, Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 74, 10807–10810 (2000)
A.S. Lipatov, S. Andreansky, R.J. Webby, D.J. Hulse, J.E. Rehg, S.S. Krause, et al., Pathogenesis of Hong Kong H5N1 influenza virus NS gene reassortants in mice: the role of cytokines and B- and T-cell responses. J. Gen. Virol. 86, 1121–1130 (2005)
H. Suzuki, R. Saito, H. Masuda, H. Oshitani, M. Sato, I. Sato, Emergence of amantadine-resistant influenza A viruses: epidemiological study. J. Infect. Chemother. 9, 195–200 (2003)
S.L. Salzberg, C. Kingsford, G. Cattoli, D.J. Spiro, D.A. Janies, M.M. Aly, et al., Genome analysis linking recent European and African influenza (H5N1) viruses. Emerg. Infect. Dis. 13(5), 713–718 (2007)
P. Puthavathana, P. Auewarakul, P.C. Charoenying, K. Sangsiriwut, P. Pooruk, K. Boonnak, et al., Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 86, 423–433 (2005)
M.N. Matrosovich, N. Zhou, Y. Kawaoka, R. Webster, The surface glycoproteins of H5 influenza viruses isolated from humans, chicken, and wild aquatic birds have distinguishable properties. J. Virol. 73, 1146–1155 (1999)
H.K. Iwatsuki, R. Kanazawa, S. Sugii, Y. Kawaoka, T. Horimoto, The index influenza A virus subtype H5N1 isolated from a human in 1997 differs in its receptor binding properties from a virulent influenza virus. J. Gen. Virol. 85, 1001–1005 (2004)
M.N. Matrosovich, T.Y. Matrosovich, T. Gray, N.A. Roberts, H.D. Klenk, Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 101, 4620–4624 (2004)
J.A. Kim, S.Y. Ryu, S.H. Seo, Cells in the respiratory and intestinal tracts of chicken have different proportions of both human and avian influenza virus receptors. J. Microbiol. 43, 366–369 (2005)
O. Werner T.C. Harder, Avian influenza. in Influenza Report 2006, ed. by B.S. Kamps C. Hoffmann W. Preiser (Flying Publisher, Paris, 2006) pp. 48–86
K. Shinya, S. Hamm, M. Hatta, H. Ito, T. Ito, Y. Kawaoka, PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320, 258–266 (2004)
Z. Li, H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li et al., Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79, 12058–12064 (2005)
W.J. Bean, S.C. Threlkeld, R.G. Webster, Biologic potential of amantadine-resistant influenza A virus in an avian model. J. Infect. Dis. 159, 1050–1056 (1989)
G. Boivin, N. Goyette, H. Bernatchez, Prolonged excretion of amantadine-resistant influenza A virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin. Infect. Dis. 34, E23–E25 (2002)
C. Sweet F.G. Hayden K.J. Jakeman S. Grambas A.J. Hay, Virulence of rimantadine-resistant human influenza A (H3N2) viruses in ferrets. J. Infect. Dis. 162 969–972 (1991)
F.G. Hayden, R.B. Belshe, R.D. Clover, A.J. Hay, M.G. Oakes, W. Soo, Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. N. Engl. J. Med. 321, 1696–1702 (1989)
WHO inter-country-consultation: influenza A/H5N1 in humans in Asia: Manila, Philippines, http://www.who.int/csr/resources/publications/influenza/WHO_CDS_CSR_GIP_2005_7/en/. Accessed 6–7 May 2005
M.A. Rameix-Welti, F. Agou, P. Buchy, S. Mardy, J.T. Aubin, M.S. Véron et al., Natural variation can significantly alter the sensitivity of influenza A (H5N1) viruses to oseltamivir. Antimicrob. Agents Chemother. 50, 3809–3815 (2006)
B. Olsen, V. Munster, A. Wallensten, J. Waldenström, A.D.M.E. Osterhaus, R.A.M. Fouchier, Global patterns of influenza A virus in wild birds. Science 312, 384–388 (2006)
Acknowledgments
The work described was supported by the Intramural Grant of the National Institute of Virology, Pune (Indian Council of Medical Research, Government of India). The authors wish to thank Dr. A. Chakrabarty, Mr. A. Walimbe, Mr. G. Prabhakar, Mr. N. Mishra, and Mr. S. Ranawade for scientific discussions, bioinformatics analyses and help in the laboratory. We are grateful to Drs. M. Chadhha and D. Mourya for their support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ray, K., Potdar, V.A., Cherian, S.S. et al. Characterization of the complete genome of influenza A (H5N1) virus isolated during the 2006 outbreak in poultry in India. Virus Genes 36, 345–353 (2008). https://doi.org/10.1007/s11262-007-0195-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-007-0195-8