Abstract
Representations of probability measures in reproducing kernel Hilbert spaces provide a flexible framework for fully nonparametric hypothesis tests of independence, which can capture any type of departure from independence, including nonlinear associations and multivariate interactions. However, these approaches come with an at least quadratic computational cost in the number of observations, which can be prohibitive in many applications. Arguably, it is exactly in such large-scale datasets that capturing any type of dependence is of interest, so striking a favourable trade-off between computational efficiency and test performance for kernel independence tests would have a direct impact on their applicability in practice. In this contribution, we provide an extensive study of the use of large-scale kernel approximations in the context of independence testing, contrasting block-based, Nyström and random Fourier feature approaches. Through a variety of synthetic data experiments, it is demonstrated that our large-scale methods give comparable performance with existing methods while using significantly less computation time and memory.
Similar content being viewed by others
1 Introduction
Given a paired sample \({\mathbf {z}} = \{ (x_i,y_i) \}^m_{i=1}\) with each \((x_i, y_i)\in {\mathcal {X}} \times {\mathcal {Y}}\) independently and identically following the joint distribution \(P_{XY}\) on some generic domains \({\mathcal {X}}\) and \({\mathcal {Y}}\), the nonparametric independence problem consists of testing whether we should reject the null hypothesis \({\mathcal {H}}_0: P_{XY} = P_X P_Y\) in favour of the general alternative hypothesis \({\mathcal {H}}_1: P_{XY} \not = P_X P_Y\), where \(P_X\) and \(P_Y\) are the marginal distributions for X and Y, respectively. This problem is fundamental and extensively studied, with wide-ranging applications in statistical inference and modelling. Classical dependence measures, such as Pearson’s product–moment correlation coefficient, Spearman’s \(\rho \), Kendall’s \(\tau \) or methods based on contingency tables are typically designed to capture only particular forms of dependence (e.g. linear or monotone). Furthermore, they are applicable only to scalar random variables or require space partitioning limiting their use to relatively low dimensions. As availability of larger datasets also facilitates building more complex models, dependence measures are sought that capture more complex dependence patterns and those that occur between multivariate and possibly high-dimensional datasets. In this light, amongst the most popular dependence measures recently have been those based on characteristic functions (Székely et al. 2007; Székely and Rizzo 2009) as well as a broader framework based on kernel methods (Gretton et al. 2005, 2008). A desirable property of consistency against any alternative—i.e. test power provably increasing to one with the sample size regardless of the form of dependence, is warranted for statistical tests based on such approaches. However, this is achieved at an expense of computational and memory requirements that increase at least quadratically with the sample size, which is prohibitive for many modern applications. Thus, a natural question is whether a favourable trade-off between computational efficiency and test power can be sought with appropriate large-scale approximations. As we demonstrate, several large-scale approximations are available in this context and they lead to strong improvements in power-per-computatonal unit performance, resulting in a fast and flexible independence testing framework responsive to all forms of dependence and applicable to large datasets.
The key quantity we consider is the Hilbert–Schmidt independence criterion (HSIC) introduced by Gretton et al. (2005). HSIC uses the distance between the kernel embeddings of probability measures in the reproducing kernel Hilbert space (RKHS) (Gretton et al. 2008; Zhang et al. 2011; Smola et al. 2007). By building on decades of research into kernel methods for machine learning (Schölkopf and Smola 2002), HSIC can be applied to multivariate observations as well as to those lying in non-Euclidean and structured domains, e.g. Gretton et al. (2008) considers independence testing on text data. HSIC has also been applied to clustering and learning taxonomies (Song et al. 2007; Blaschko and Gretton 2009), feature selection (Song et al. 2012), causal inference (Peters et al. 2014; Flaxman et al. 2015; Zaremba and Aste 2014) and computational linguistics (Nguyen and Eisenstein 2016). A closely related dependence coefficient that measures all types of dependence between two random vectors of arbitrary dimensions is the distance covariance (dCov) of Székely et al. (2007), Székely and Rizzo (2009), which measures distances between empirical characteristic functions or equivalently measures covariances with respect to a stochastic process (Székely and Rizzo 2009), and its normalised counterpart, distance correlation (dCor). RKHS-based dependence measures like HSIC are in fact extensions of dCov—Sejdinovic et al. (2013b) shows that dCov can be understood as a form of HSIC with a particular choice of kernel. Moreover, dCor can be viewed as an instance of kernel matrix alignment of Cortes et al. (2012). As we will see, statistical tests based on estimation of HSIC and dCov are computationally expensive and require at least \({\mathcal {O}}(m^2)\) time and storage complexity, where m is the number of observations, just to compute an HSIC estimator which serves as a test statistic. In addition, the complicated form of the asymptotic null distribution of the test statistics necessitates either permutation testing (Arcones and Gine 1992) (further increasing the computational cost) or even more costly direct sampling from the null distribution, requiring eigendecompositions of kernel matrices using the spectral test of Gretton et al. (2009), with a cost of \({\mathcal {O}}(m^3)\).Footnote 1 These memory and time requirements often make the HSIC-based tests infeasible for practitioners.
In this paper, we consider several ways to speed up the computation in HSIC-based tests. More specifically, we introduce three fast estimators of HSIC: the block-based estimator, the Nyström estimator and the random Fourier feature (RFF) estimator and study the resulting independence tests. In the block-based setting, we obtain a simpler asymptotic null distribution as a consequence of the central limit theorem in which only asymptotic variance needs to be estimated—we discuss possible approaches for this. RFF and Nyström estimators correspond to the primal finite-dimensional approximations of the kernel functions and as such also warrant estimation of the null distribution in linear time—we introduce a novel spectral tests based on eigendecompositions of primal covariance matrices, which avoid permutation approach and significantly reduce the computational expense for the direct sampling from the null distribution.
1.1 Related work
Some of the approximation methods considered in this paper were inspired by their use in a related context of two-sample testing. In particular, the block-based approach for two-sample testing was studied in Gretton et al. (2012b, 2012a), Zaremba et al. (2013) under the name of linear time MMD (maximum mean discrepancy), i.e. the distance between the mean embeddings of the probability distributions in the RKHS. The approach estimates MMD on a small block of data and then averages the estimates over blocks to obtain the final test statistic. Our block-based estimator of HSIC follows exactly the same strategy. On the other hand, The Nyström method (Williams and Seeger 2001; Snelson and Ghahramani 2006) is a classical low-rank kernel approximation technique, where data are projected into lower-dimensional subspaces of RKHS (spanned by so-called inducing variables). Such an idea is popular in fitting sparse approximations to Gaussian process (GP) regression models, allowing reduction in the computational cost from \({\mathcal {O}}(m^3)\) to \({\mathcal {O}}(n^2m)\) where \(n \ll m\) is the number of inducing variables. To the best of our knowledge, Nyström approximation was not studied in the context of hypothesis testing. Random Fourier feature (RFF) approximations (Rahimi and Recht 2007), however, due to their relationship with evaluations of empirical characteristic functions, do have a rich history in the context of statistical testing—as discussed in Chwialkowski et al. (2015), which also proposes an approach to scale up kernel-based two-sample tests by additional smoothing of characteristic functions, thereby improving the test power and its theoretical properties. Moreover, the approximation strategy of MMD and two-sample testing through primal representation using RFF have also been studied in Zhao and Meng (2015), Sutherland and Schneider (2015), Lopez-Paz (2016). In addition, Lopez-Paz et al. (2013) first proposed the idea of applying RFF in order to construct an approximation to a kernel-based dependence measure. More specifically, they develop randomised canonical correlation analysis (RCCA) (see also Lopez-Paz 2014, 2016) approximating the nonlinear kernel-based generalisation of the canonical correlation analysis (Lai and Fyfe 2000; Bach and Jordan 2002) and using a further copula transformation, construct a test statistic termed RDC (randomised dependence coefficient) requiring \(O(m\log m)\) time to compute. Under suitable assumptions, Bartlett’s approximation (Mardia et al. 1979) provides a closed form expression for the asymptotic null distribution of this statistic which further results in a distribution-free test, leading to an attractive option for large-scale independence testing. We extend these ideas based on RFF to construct approximations of HSIC and dCov/dCor, which are conceptually distinct kernel-based dependence measures from that of kernel CCA, i.e. they measure different types of norms of RKHS operators (operator norm vs Hilbert–Schmidt norm).
In fact, the Nyström and RFF approximations can also be viewed through the lense of nonlinear canonical analysis framework introduced by Dauxois and Nkiet (1998). This is the earliest example we know where nonlinear dependence measures based on spectra of appropriate Hilbert space operators are studied. In particular, the cross-correlation operator with respect to a dictionary of basis functions in \(L_2\) (e.g. B-splines) is considered in Dauxois and Nkiet (1998). Huang et al. (2009) links this framework to the RKHS perspective. The functions of the spectra that were considered in Dauxois and Nkiet (1998) are very general, but the simplest one (sum of the squared singular values) can be recast as the normalised cross-covariance operator (NOCCO) of Fukumizu et al. (2008), which considers the Hilbert–Schmidt norm of the cross-correlation operator on RKHSs and as such extends kernel CCA to consider the entire spectrum. While in this work we focus on HSIC (Hilbert–Schmidt norm of the cross-covariance operator), which is arguably the most popular kernel dependence measure in the literature, a similar Nyström or RFF approximation can be applied to NOCCO as well—we leave this as a topic for future work.
The paper is structured as follows: in Sect. 2, we first provide some necessary definitions from the RKHS theory and review the aforementioned Hilbert–Schmidt independence criterion (HSIC) and discuss its biased and unbiased quadratic time estimators. Then, Sect. 2.3 gives the asymptotic null distributions of estimators (proofs provided in Section A). In Sect. 3, we develop a block-based HSIC estimator and derive its asymptotic null distribution. Following this, a linear time asymptotic variance estimation approach is proposed. In Sects. 4.1 and 4.2, we propose Nyström HSIC and RFF HSIC estimator, respectively, both with the corresponding linear time null distribution estimation approaches. Finally, in Sect. 5, we explore the performance of the three testing approaches on a variety of challenging synthetic data.
2 Background
This section starts with a brief overview of the key concepts and notation required to understand the RKHS theory and kernel embeddings of probability distributions into the RKHS. It then provides the definition of HSIC which will serve as a basis for later independence tests. We review the quadratic time biased and unbiased estimators of HSIC as well as their respective asymptotic null distributions. As the final part of this section, we outline the construction of independence tests in quadratic time.
2.1 RKHS and embeddings of measures
Let \(\mathcal {Z }\) be any topological space on which Borel measures can be defined. By \(\mathcal {M(Z)}\) we denote the set of all finite-signed Borel measures on \({\mathcal {Z}}\) and by \({\mathcal {M}}^1_+ ({\mathcal {Z}})\) the set of all Borel probability measures on \({\mathcal {Z}}\). We will now review the basic concepts of RKHS and kernel embeddings of probability measures. For further details, see Berlinet and Thomas-Agnan (2004), Steinwart and Christmann (2008), Sriperumbudur (2010).
Definition 1
Let \({\mathcal {H}}\) be a Hilbert space of real-valued function defined on \({\mathcal {Z}}\). A function \(k:{\mathcal {Z}} \times {\mathcal {Z}} \rightarrow {\mathbb {R}}\) is called a reproducing kernel of \({\mathcal {H}}\) if:
-
1.
\(\forall z \in {\mathcal {Z}}, k(\cdot ,z) \in {\mathcal {H}}\)
-
2.
\(\forall z \in {\mathcal {Z}}, \forall f \in {\mathcal {H}}, \langle f,k(\cdot ,z) \rangle _{\mathcal {H}} = f(z).\)
If \({\mathcal {H}}\) has a reproducing kernel, it is called a reproducing kernel Hilbert space (RKHS).
As a direct consequence, for any \(x,y \in {\mathcal {Z}}\),
In machine-learning literature, a notion of kernel is understood as an inner product between feature maps (Steinwart and Christmann 2008). By (1), every reproducing kernel is a kernel in this sense, corresponding to a canonical feature map \(x\mapsto k(\cdot ,x)\).
For \(x,y \in {\mathbb {R}}^p\), some examples of reproducing kernels are
-
Linear kernel: \(k(x,y) = x^T y\);
-
Polynomial kernel of degree \(d \in {\mathbb {N}}\): \(k(x,y) = (x^T y + 1)^d \);
-
Gaussian kernel with bandwidth \(\sigma > 0\): \(k(x,y) = \exp (-\frac{\Vert x- y\Vert ^2}{2\sigma ^2})\);
-
Fractional Brownian motion covariance kernel with parameter \(h\in (0,1)\): \(k(x,y)=\frac{1}{2}\left( \Vert x\Vert ^{2h}+\Vert y\Vert ^{2h}-\,\Vert x-y\Vert ^{2h}\right) \)
Checking whether a given function k is a valid reproducing kernel can be onerous. Fortunately, the Moore–Aronszajn theorem (Aronszajn 1950) gives a simple characterisation: for any symmetric, positive-definite function \(k: {\mathcal {Z}} \times {\mathcal {Z}} \rightarrow {\mathbb {R}}\), there exists a unique Hilbert space of functions \({\mathcal {H}}\) defined on \({\mathcal {Z}}\) such that k is reproducing kernel of \({\mathcal {H}}\) (Berlinet and Thomas-Agnan 2004). RKHS are precisely the space of functions where norm convergence implies pointwise convergence and are as a consequence relatively well behaved comparing to other Hilbert spaces. In nonparametric testing, as we consider here, a particularly useful setup will be representing probability distributions and, more broadly, finite-signed Borel measures \(\nu \in \mathcal {M(Z)}\) with elements of an RKHS (Smola et al. 2007).
Definition 2
Let k be a kernel on \({\mathcal {Z}}\), and \(\nu \in \mathcal {M(Z)}\). The kernel embedding of measure \(\nu \) into the RKHS \({\mathcal {H}}_k\) is \(\mu _k (\nu ) \in {\mathcal {H}}_k\) such that
It is understood from this definition that the integral of any RKHS function f under the measure \(\nu \) can be evaluated as the inner product between f and the kernel embedding \(\mu _k (\nu )\) in the RKHS \( {\mathcal {H}}_k.\) As an alternative, the kernel embedding can be defined through the use of Bochner integral \(\mu _k (\nu ) = \int k(\cdot ,z) \mathrm{d}\nu (z)\). Any probability measure is mapped to the corresponding expectation of the canonical feature map. By Cauchy–Schwarz inequality and the Riesz representation theorem, a sufficient condition for the existence of an embedding of \(\nu \) is that \(\nu \in {\mathcal {M}}^{1/2}_{k}({\mathcal {Z}})\), where we adopt notation from Sejdinovic et al. (2013b): \({\mathcal {M}}^\theta _{k}({\mathcal {Z}}) = \left\{ \nu \in {\mathcal {M}}(Z): \int k^\theta (z,z) \mathrm{d}|\nu |(z) < \infty \right\} \), which is, e.g. satisfied for all finite measures if k is a bounded function (such as Gaussian kernel).
Embeddings allow measuring distances between probability measures, giving rise to the notion of maximum mean discrepancy (MMD) (Borgwardt et al. 2006; Gretton et al. 2012b).
Definition 3
Let k be a kernel on \({\mathcal {Z}}\). The squared distance between the kernel embeddings of two probability measures P and Q in the RKHS,
is called maximum mean discrepancy (MMD) between P and Q with respect to k.
When the corresponding kernels are characteristic (Sriperumbudur 2010), embedding is injective and MMD is a metric on probability measures. The estimators of MMD are useful statistics in nonparametric two-sample testing (Gretton et al. 2012b), i.e. testing if two given samples are drawn from the same probability distribution. For any kernels \(k_{{\mathcal {X}}}\) and \(k_{{\mathcal {Y}}}\) on the respective domains \({\mathcal {X}}\) and \({\mathcal {Y}}\), it is easy to check that \(k=k_{{\mathcal {X}}}\otimes k_{{\mathcal {Y}}}\) given by
is a valid kernel on the product domain \({\mathcal {X}} \times {\mathcal {Y}}\). Its canonical feature map is \((x,y)\mapsto k_{{\mathcal {X}}}(\cdot ,x)\otimes k_{{\mathcal {Y}}}(\cdot ,y)\) where \(\varphi _{x,y}=k_{{\mathcal {X}}}(\cdot ,x)\otimes k_{{\mathcal {Y}}}(\cdot ,y)\) is understood as a function on \({\mathcal {X}} \times {\mathcal {Y}}\), i.e. \(\varphi _{x,y}(x',y')=k_{{\mathcal {X}}}(x',x)k_{{\mathcal {Y}}}(y',y)\). The RKHS of \(k=k_{{\mathcal {X}}}\otimes k_{{\mathcal {Y}}}\) is in fact isometric to \({\mathcal {H}}_{k_{\mathcal {X}}}\otimes {\mathcal {H}}_{k_{\mathcal {Y}}}\), which can be viewed as the space of Hilbert–Schmidt operators between \({\mathcal {H}}_{k_{\mathcal {Y}}}\) and \({\mathcal {H}}_{k_{\mathcal {X}}}\) (Lemma 4.6 of Steinwart and Christmann (2008)). We are now ready to define an RKHS-based measure of dependence between random variables X and Y.
Definition 4
Let X and Y be random variables on domains \({\mathcal {X}}\) and \({\mathcal {Y}}\) (non-empty topological spaces). Let \(k_{{\mathcal {X}}}\) and \(k_{{\mathcal {Y}}}\) be kernels on \({\mathcal {X}}\) and \({\mathcal {Y}}\) respectively. Hilbert–Schmidt independence criterion (HSIC) \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}(X,Y)\) of X and Y is MMD between the joint measure \(P_{XY}\) and the product of marginals \(P_{X}P_{Y}\), computed with the product kernel \(k=k_{{\mathcal {X}}}\otimes k_{{\mathcal {Y}}}\), i.e.
HSIC is well defined whenever \(P_X \in {\mathcal {M}}^1_{k_{{\mathcal {X}}}}({\mathcal {X}})\) and \(P_Y \in {\mathcal {M}}^1_{k_{{\mathcal {Y}}}}({\mathcal {Y}})\) as this implies \(P_{XY} \in {\mathcal {M}}^{1/2}_{k_{{\mathcal {X}}}\otimes k_{{\mathcal {Y}}}}({\mathcal {X}} \times {\mathcal {Y}})\) (Sejdinovic et al. 2013b). The name of HSIC comes from the operator view of the RKHS \({\mathcal {H}}_{k_{\mathcal {X}}\otimes k_{\mathcal {Y}}}\). Namely, the difference between embeddings \({\mathbb {E}}_{XY}[k_{\mathcal {X}}(.,X) \otimes k_{\mathcal {Y}}(.,Y)] - {\mathbb {E}}_X k_{\mathcal {X}}(.,X) \otimes {\mathbb {E}}_Y k_{\mathcal {Y}}(.,Y)\) can be identified with the cross-covariance operator \(C_{XY}:{\mathcal {H}}_{k_{\mathcal {Y}}}\rightarrow {\mathcal {H}}_{k_{\mathcal {X}}}\) for which \(\langle f,C_{XY}g\rangle _{{\mathcal {H}}_{k_{\mathcal {X}}}}={\text {Cov}}\left[ f(X)g(Y)\right] \), \(\forall f\in {\mathcal {H}}_{k_{\mathcal {X}}},g\in {\mathcal {H}}_{k_{\mathcal {Y}}}\) (Gretton et al. 2005, 2008). HSIC is then simply the squared Hilbert–Schmidt norm \(\Vert C_{XY}\Vert _{HS}^2\) of this operator, while distance correlation (dCor) of Székely et al. (2007), Székely and Rizzo (2009) can be cast as \(\Vert C_{XY}\Vert _{HS}^2 / \Vert C_{XX}\Vert _{HS}\Vert C_{YY}\Vert _{HS}\) (Sejdinovic et al. 2013b, Appendix A). In the sequel, we will suppress dependence on kernels \(k_{\mathcal {X}}\) and \(k_{\mathcal {Y}}\) in notation \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}(X,Y)\) where there is no ambiguity.
Repeated application of the reproducing property gives the following equivalent representation of HSIC (Smola et al. 2007):
Proposition 1
The HSIC of X and Y can be written as:
2.2 Estimation of HSIC
Using the form of HSIC in (6), given an iid sample of z = \(\{ (x_i, y_i) \}^m_{i=1}\) from the joint distribution \(P_{XY}\), an unbiased estimator of HSIC can be obtained as a sum of three U-statistics (Gretton et al. 2008):
where the index set \({\mathbf {i}}^m_r\) denotes the set of all r-tuples drawn without replacement from \(\{ 1, \ldots , m \}\), \((K_x)_{ij} := k_{{\mathcal {X}}}(x_i,x_j)\) and \((K_y)_{ij} := k_{{\mathcal {Y}}}(y_i,y_j)\). Naïve computation of (7) would require \({\mathcal {O}}(m^4)\) operations. However, an equivalent form which needs \({\mathcal {O}}(m^2)\) operations is given in Song et al. (2012) as
where \(\tilde{K}_x = K_x - diag(K_x)\) (i.e. the kernel matrix with diagonal elements set to zero) and similarly for \(\tilde{K}_y\). \({\mathbbm {1}}\) is a vector of 1s of relevant dimension.
We will refer to the above as the quadratic time estimator. Gretton et al. (2008) note that the V-statistic estimator (or the quadratic time-biased estimator) of HSIC can be an easier-to-use alternative for the purposes of independence testing, since the bias is accounted for in the asymptotic null distribution. The V-statistic is given by
where the summation indices are now drawn with replacement. Further, it can be simplified as follows to reduce the computation:
where \(H = I_m- \frac{1}{m}{\mathbbm {1}}{\mathbbm {1}}^T\) is an \(m \times m\) centering matrix. (9) gives an intuitive understanding of the HSIC statistic: it measures average similarity between the centred kernel matrices, which are in turn similarity patterns within the samples.Footnote 2
2.3 Asymptotic null distribution of estimators
The asymptotic null distribution of the biased HSIC statistic defined in (9) computed using a given dataset converges in distribution in Theorem 1 below. This asymptotic distribution builds the theoretical foundation for the spectral testing approach (described in Sect. 2.4.2) that we will use throughout the paper.
Theorem 1
(Asymptotic null distribution of the biased HSIC) Under the null hypothesis, let the dataset \({\mathbf {z}} = \{(x_i,y_i)\}^m_{i=1} \overset{i.i.d.}{\sim }P_{XY}=P_XP_Y\), with \(P_X \in {\mathcal {M}}^2_{k_{{\mathcal {X}}}}({\mathcal {X}})\) and \(P_Y \in {\mathcal {M}}^2_{k_{{\mathcal {Y}}}}({\mathcal {Y}})\), then
where \(N_{i,j} \overset{i.i.d.}{\sim }{\mathcal {N}}\)(0,1), \( \forall \ i,j \in {\mathbb {N}}\) and \(\{\lambda _i\}^\infty _{i=1}\), \(\{\eta _j\}^\infty _{j=1}\) are the eigenvalues of the integral kernel operators \(S_{\tilde{k}_{P_x}}\) and \(S_{\tilde{k}_{P_y}}\), where the integral kernel operator \(S_{\tilde{k}_{P}}: L^2_P({\mathcal {Z}}) \rightarrow L^2_P({\mathcal {Z}})\) is given by
where \(\tilde{k}_P(z, z')\) is the kernel centred at probability measure P:
with \(W,W'\overset{i.i.d.}{\sim }P.\)
For completeness, the proof of this theorem, which is a consequence of Lyons (2013, Theorem 2.7) and the equivalence between distance-based and RKHS-based independence statistics (Sejdinovic et al. 2013b) is given in Appendix. As remarked by Sejdinovic et al. (2013b), the finite marginal moment conditions imply that the integral operators \(S_{\tilde{k}_{{\mathcal {X}}}}\) and \(S_{\tilde{k}_{{\mathcal {Y}}}}\) are trace class and hence Hilbert–Schmidt (Reed and Simon 1980). Anderson et al. noted that the form of the asymptotic distribution of the V statistics requires the integral operators being trace class but that of the U statistics only requires them being Hilbert–Schmidt (Anderson et al. 1994; Sejdinovic et al. 2013b). Using the same notation as in the case of the V-statistic, the asymptotic distribution of the U-statistic in (7) can be written as:
under the null hypothesis.
We note that Chwialkowski and Gretton (2014) (Lemma 2 and Theorem 1) proves a more general result, applicable to dependent observations under certain mixing conditions where the i.i.d. setting is a special case. Moreover, Rubenstein et al. (2015) (Theorem 5 and 6) provides another elegant proof in the context of three-variable interaction testing from Sejdinovic et al. (2013a). However, both Chwialkowski and Gretton (2014) and Rubenstein et al. (2015) assume boundedness of \(k_{\mathcal {X}}\) and \(k_{\mathcal {Y}}\), while our proof in Appendix assumes a weaker condition of finite second moments for both \(k_{\mathcal {X}}\) and \(k_{\mathcal {Y}}\), thus making the result applicable to unbounded kernels such as the Brownian motion covariance kernel.
Under the alternative hypothesis that \(P_X P_Y \not = P_{XY}\), Gretton et al. (2008) remarked that \(m \varXi _{b,k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {Z}})\) converges to HSIC with the corresponding appropriately centred and scaled Gaussian distribution as \(m \rightarrow \infty \):
where the variance \(\sigma ^2_u = 16({\mathbb {E}}_{z_i} ({\mathbb {E}}_{z_j,z_q,z_r}(h_{ijqr}))^2 -\hbox {HSIC})\) and \(h_{ijqr}\) is defined as
with all ordered quadruples (t, u, v, w) drawn without replacement from (i, j, q, r) and assuming \({\mathbb {E}} (h^2) < \infty \). In fact, under the alternative hypothesis, the difference between \(m \varXi _{b}({\mathbf {Z}})\) (i.e. the V-statistic) and the U-statistic drops as 1 / m and hence asymptotically the two statistics converges to the same distribution (Gretton et al. 2008).
2.4 Quadratic time null distribution estimations
We would like to design independence tests with an asymptotic Type I error of \(\alpha \) and hence we need an estimate of the \((1-\alpha )\) quantile of the null distribution. Here, we consider two frequently used approaches, namely the permutation approach and the spectral approach, that require at least quadratic time both in terms of memory and computation time. The biased V-statistic will be used because of its neat and compact formulation.
2.4.1 Permutation approach
Consider an iid sample \({\mathbf {z}} = \left\{ (x_i,y_i)\right\} ^m_{i=1}\) with chosen kernels \(k_{{\mathcal {X}}}\) and \(k_{{\mathcal {Y}}}\), respectively, the permutation/bootstrap approach Arcones and Gine (1992) proceed in the following manner. Suppose the total number of shuffles is fixed at \(N_p\), we first compute \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z}})\) using \({\mathbf {z}}\), \(k_{{\mathcal {X}}}\) and \(k_{{\mathcal {Y}}}\). Then, for each shuffle, we fix the \(\{ x_i \}^m_{i=1}\) and randomly permute the \(\{ y_i \}^m_{i=1}\) to obtain \({\mathbf {z}}^* = \left\{ (x_i,y^*_i)\right\} ^m_{i=1}\) and subsequently compute \(\varXi ^*_{k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z^*}})\). The one-sided p-value in this instance is the proportion of HSIC values computed on the permuted data that are greater than or equal to \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z}})\).
The computational time is \({\mathcal {O}}\)(number of shuffles \(\times m^2)\) for this approach, where the number of shuffles determines the extend to which we have explored the sampling distribution. In other words, a small number of shuffles means that we may only obtained realisations from the mode of the distribution and hence the tail structure is not adequately captured. Although a larger number of shuffles ensures the proper exploration of the sampling distribution, the computation cost can be high.
2.4.2 Spectral approach
Gretton et al. have shown (Theorem 1 Gretton et al. 2009) that the empirical finite sample estimate of the null distribution converges in distribution to its population counterpart provided that the eigenspectrums \(\{ \gamma _r \}^\infty _{r=1}\) of the integral operator \(S_{\tilde{k} }\): \(L^2_\theta ({\mathcal {X}} \times {\mathcal {Y}}) \rightarrow L^2_\theta ({\mathcal {X}} \times {\mathcal {Y}})\) is square root summable, i.e.
Note, the integral operator \(S_{\tilde{k} }\) is the tensor product of the operators \(S_{\tilde{k}_{{\mathcal {X}}} } \) and \(S_{\tilde{k}_{{\mathcal {X}}} } \):
and the eigenvalues of this operator is hence the product of the eigenvalues of these two operators.
The spectral approach (Gretton et al. 2009; Zhang et al. 2011) requires that we first calculate the centred Gram matrices \(\widetilde{K}_X = HK_{X}H\) and \(\widetilde{K}_Y = HK_{Y}H\) for the chosen kernel \(k_{{\mathcal {X}}}\) and \(k_{{\mathcal {Y}}}\). Then, we compute the \(m \varXi _{b,k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z}})\) statistics according to (9). Next, the spectrums (i.e. eigenvalues) \(\{\lambda _i\}^m_{i=1}\) and \(\{\eta _i\}^m_{i=1}\) of \(\widetilde{K}_X\) and \(\widetilde{K}_Y\) are, respectively, calculated. The empirical null distribution can be simulated by simulating a large enough i.i.d samples from the standard Normal distribution (Zhang et al. 2011) and then generate the test statistic according to (10). Finally, the p value is computed by calculating the proportion of simulated samples that are greater than or equal to the observed \(m \varXi _{b,k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z}})\) value.
Additionally, Zhang et al. (2011) has provided an approximation to the null distribution with a two-parameter Gamma distribution. Despite the computational advantage of such an approach, the permutation and spectral approaches are still preferred since there is no consistency guarantee for the Gamma distribution approach.
3 Block-based HSIC
The quadratic time test statistics are prohibitive for large dataset as it requires \({\mathcal {O}}(m^2)\) time in terms of storage and computation. Furthermore, one requires an approximation of the asymptotic null distribution in order to compute the p value. As we discussed in the previous section, this is usually done by randomly permute the Y observations (i.e. the permutation approach) or by performing an eigendecomposition of the centred kernel matrices for X and Y (i.e. the spectral approach). Both approaches are expensive in terms of memory and can be computationally infeasible. In this section, we propose a block-based estimator of HSIC which reduce the computational time to linear in the number of samples. The asymptotic null distribution of this estimator will be shown to have a simple form as a result of the central limit theorem (CLT).
3.1 The block HSIC statistic
Let us consider that the sample is split into blocks of size \(B\ll m\): \(\left\{ x_{i},y_{i}\right\} _{i=1}^{m} \overset{i.i.d.}{\sim }P_{XY}\) becomes \(\{ \{ x_{i}^{(b)}, y_{i}^{(b)} \} _{i=1}^{B}\} _{b=1}^{m/B}\) (where we assumed for simplicity that m is divisible by B). We follow the approach from Zaremba et al. (2013), Sejdinovic et al. (2014) and extend it to independence testing. We compute the unbiased HSIC statistics (Eq. 7) on each block \(b \in \{ 1, \ldots , \frac{m}{B} \}\):
and average them over blocks to establish the block-based estimator for HSIC:
3.2 Null distribution of block-based HSIC
For the block HSIC statistic, the asymptotic null distribution is a consequence of the central limit theorem (CLT) under the regime where \(m \rightarrow \infty \), \(B \rightarrow \infty \) and \(\frac{m}{B} \rightarrow \infty \).Footnote 3 First of all, note that the linear time test statistic \(\hat{\eta }_k\) is an average of block-based statistics \(\hat{\eta }_b\) for \(b \in \{1, \ldots , \frac{m}{B}\}\) which are independent and identically distributed. Secondly, we recall that \({\mathbb {E}}(\hat{\eta }_b) = 0\) for \(\hat{\eta }_b\) is an unbiased estimator of HSIC. Finally, \({\mathbb {V}}ar(\hat{\eta }_k) = \frac{B}{m} {\mathbb {V}}ar(\hat{\eta }_b) = \frac{B}{m}\frac{1}{B^2} {\mathbb {V}}ar(W)\) with W being the random variable distributed according to \(\sum ^\infty _{i=1} \sum ^\infty _{j=1} \lambda _i \eta _j (N_{i,j}^2-1)\). In the limit as \(m \rightarrow \infty \), \(B \rightarrow \infty \) and \(\frac{m}{B} \rightarrow \infty \):
where the variance \(\sigma ^2_{k,0}\) is the variance of the null distributions in Expression (10) and (13), i.e. the variance of W and it is given by
3.3 Linear time null distribution estimation
Expression (18) guarantees the Gaussianity of the null distribution of the block-based statistic and, henceforth, makes the computation of p-value straightforward. We simply return the test statistic \(\sqrt{mB}\frac{ \hat{\eta }_{b}}{\sqrt{\hat{\sigma }^2_{k,0}}}\) and compare against the corresponding quantile of \({\mathcal {N}}(0,1 )\) which is the approach taken in Gretton et al. (2012a), Zaremba et al. (2013), Sejdinovic et al. (2014). Note that the resulting null distribution is actually a t-distribution but with a very large number of degrees of freedom, which can be treated as a Gaussian distribution.
The difficulty of estimating the null distribution lies in estimating \(\sigma ^2_{k,0}\). We suggest two ways to estimate such variance (Sejdinovic et al. 2014): within-block permutation and within-block direct estimation. These two approaches are at most quadratic in B within each block which means that the computational cost of estimating the variance is of the same order as that of computing the statistic itself.
Within-block permutation can be done as follows. Within each block, we compute the test statistic using (16). At the same time, we track in parallel a sequence \(\hat{\eta }^*_{b}\) obtained using the same formula but with \(\{y_i\}^m_{i=1}\) underwent a permutation. The former is used to calculate the overall block statistics and the latter is used to estimate the null variance \(\hat{\sigma }^2_{k,0} = B^2 {\mathbb {V}}ar [ \{\hat{\eta }^*_{b} \}^{m/B}_{b=1} ]\) as the independence between the samples holds by construction.
Within-block direct estimation can be achieved by using (20) and the corresponding unbiased estimates of \({\mathbb {E}}_{XX'}(\tilde{k}^2_{P_X}(X,X'))\) and \({\mathbb {E}}_{YY'}(\tilde{k}^2_{P_Y}(Y,Y'))\) which can be calculated as follows. For X, the estimate of the variance for each block is given by Song et al. (2012):
Then, we compute
to obtain an unbiased estimate for \({\mathbb {E}}_{XX'}(\tilde{k}^2_{P_X}(X,X'))\). Similarly, replacing all x with y, we obtain an unbiased estimate for \({\mathbb {E}}_{YY'}(\tilde{k}^2_{P_Y}(Y,Y'))\). The estimate of the variance is therefore:
Such block-based approach is trivially parallelisable: the HSIC statistic defined in (16) as well as the variance estimator of (21) can be computed separately for each block. This may provide further computational efficiency gains. As remarked in Sejdinovic et al. (2014), we note that under the null hypothesis, the approach undertaken by Zaremba et al. (2013) is to estimate the null variance directly with the empirical variance of \(\left\{ \hat{\eta }_{b}\right\} _{b=1}^{m/B}\). As the null variance is consistently estimated under the null hypothesis, this ensures the correct level of Type I error. However, without using the variance of the “bootstrap samples”, such an estimate of the null variance will systematically overestimate as B grows with m. Hence, it will result in a reduced statistical power due to inflated p values.
Regarding the choice of B, Zaremba et al. (2013) discussed that the null distribution is close to that guaranteed by the CLT when B is small, and hence, the Type I error will be closer to the desired level. However, the disadvantage is the small statistical power for each given sample size. Conversely, Zaremba et al. (2013) pointed out that a larger B results in a lower variance empirical null distribution and hence higher power. Hence, they suggested a sensible family of heuristics is to set \(B=\lfloor m^{\gamma }\rfloor \) for some \( 0<\gamma <1\). As a result, the complexity of the block-based test is \({\mathcal {O}}(Bm) = {\mathcal {O}}(m^{1+\gamma })\).
4 Approximate HSIC through primal representations
Having discussed how we can construct a linear time HSIC test by processing the dataset in blocks, we now move on to consider how the scaling up can be done through low-rank approximations of the Gram matrix. In particular, we will discuss Nyström type approximation (Sect. 4.1) and random Fourier features (RFF) type approximation (Sect. 4.2). Both types of approximation act directly on the primal representation of the kernel hence provide finite representations of the feature maps.
Recall that the definition of HSIC of X and Y in (6) can also be written in terms of the cross-covariance operator \(C_{XY}\): \( \varXi (X,Y) = \Vert C_{XY} \Vert ^2_{{\mathcal {H}}_{k_{\mathcal {X}}} \otimes {\mathcal {H}}_{k_{\mathcal {Y}}}}\) (Gretton et al. 2005, 2008). Given a data set \({\mathbf {z}} = \{ (x_i,y_i) \}^m_{i=1}\) with \(x_i \in {\mathbb {R}}^{D_x}\) and \(y_i \in {\mathbb {R}}^{D_y}\) for all i, consider the empirical version of \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}(X,Y)\) with kernels \(k_{\mathcal {X}}\) and \(k_{\mathcal {Y}}\) for X and Y, respectively:
where the Hilbert–Schmidt norm is taken in the product space \({{\mathcal {H}}_{k_{\mathcal {X}}} \otimes {\mathcal {H}}_{k_{\mathcal {Y}}}} \). Note, this empirical cross-covariance operator is infinite dimensional. However, when approximate feature representations are used, the cross-covariance operator is a finite-dimensional matrix and hence the Hilbert–Schmidt norm is equivalent to the Frobenius norm (F).
If we let \(\bar{\phi }(x_i) = k_{\mathcal {X}}(\cdot ,x_i)- \frac{1}{m} \sum ^m_{r=1} k_{\mathcal {X}}(\cdot ,x_r)\) and \(\bar{\psi }(y_i) = k_{\mathcal {Y}}(\cdot ,y_i)- \frac{1}{m} \sum ^m_{r=1} k_{\mathcal {Y}}(\cdot ,y_r)\), the above expression can be further simplified as
Hence, we obtain the expression in (9). If instead, we replace \(\bar{\phi }(x_i)\) and \(\bar{\psi }(y_i)\) by the corresponding low-rank approximations \(\tilde{\phi }(x_i) = \tilde{k}_{\mathcal {X}}(\cdot ,x_i)- \frac{1}{m} \sum ^m_{r=1} \tilde{k}_{\mathcal {X}}(\cdot ,x_r)\) and \(\tilde{\psi }(y_i) = \tilde{k}_{\mathcal {Y}}(\cdot ,y_i)- \frac{1}{m} \sum ^m_{r=1} \tilde{k}_{\mathcal {Y}}(\cdot ,y_r)\), we can obtain the approximated HSIC statistics. The details of which are provided in the following sections.
4.1 Nyström HSIC
In this section, we use the traditional Nyström approach to provide an approximation that consider the similarities between the so-called inducing variables and the given dataset. We will start with a review of Nyström method and then we will provide the explicit feature map representation of the Nyström HSIC estimator. To finish, we will discuss two null distribution estimation approaches that cost at most linear in the number of samples.
The reduced-rank approximation matrix provided by Nyström method (Williams and Seeger 2001) represents each data point by a vector based on its kernel similarity to the inducing variables and the induced kernel matrix. The approximation is achieved by randomly sample n data points (i.e. inducing variables) from the given m samples and compute the approximate kernel matrix \(\tilde{K} \approx K\) as:
where each of \(K_{r,s}\) can be think of as the \(r \times s\) block of the full Gram matrix K computed using all given samples.
Further, we can write Eq. (29) as:
Hence, an explicit feature representation of \(\tilde{K}\) is obtained. Note that Snelson and Ghahramani (2006) further relaxed the setting and propose to use inducing points that are not necessarily a subset of the given data but only need to explain the dataset well for a good performance.
4.1.1 The Nyström HSIC statistic
To further reduce computation cost, we propose to approximate this reduced-rank kernel matrix \(\tilde{K}\) with the uncentered covariance matrix \(\tilde{C}\) that is \(n \times n\):
Let us denote \(\tilde{C}_X = \tilde{\varPhi }_X^T \tilde{\varPhi }_X \) and \(\tilde{C}_Y = \tilde{\varPhi }_Y^T \tilde{\varPhi }_Y \). In order to approximate the biased HSIC estimator (Eq. 9) using this explicit feature map representation, the \(\tilde{\varPhi }_X\) and \(\tilde{\varPhi }_Y\) needed to be centred. We suggest centre each column separately by subtracting its mean for both \(\tilde{\varPhi }_X\) and \(\tilde{\varPhi }_Y\), i.e. denote \( \hat{\varPhi }_.=(I_m - \frac{1}{m}{\mathbbm {1}}{\mathbbm {1}}^T)\tilde{\varPhi }_. = H \tilde{\varPhi }. \in {\mathcal {R}}^{m \times n_.}\) for X and Y, respectively.
Using the methods described above, we can substitute approximated kernel functions \(\hat{k}_{{\mathcal {X}}} = \hat{\varPhi }_X\) and \(\hat{k}_{{\mathcal {Y}}} = \hat{\varPhi }_Y\) into the empirical version of \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}(X,Y)\) (24):
where \(\tilde{\varPhi }_X(x_i) \in {\mathbb {R}}^{n_x}\) and \(\tilde{\varPhi }_Y(y_i) \in {\mathbb {R}}^{n_y}\) can both be computed in linear time in m. This is the biased Nyström estimator of HSIC. Essentially, we approximate the cross-covariance operator \(C_{XY}\) by the Nyström estimator \(\tilde{C}_{XY} = \frac{1}{m} \tilde{\varPhi }_X^T \tilde{\varPhi }_Y \in {\mathbb {R}}^{n_x \times n_y}\) which only requires \({\mathcal {O}}(n_x n_y m )\). In essence, the HSIC statistic computed using Nyström as we described here is a HSIC statistic computed using a different kernel. As a remark, we note that it is not immediately clear how one can choose the inducing points optimally. For the synthetic data experiments in Sect. 5, we simulate the inducing data from the same distribution as data X. But, we will leave the more general case as further work.
4.1.2 Null distribution estimations
Having introduced the biased Nyström HSIC statistics, we will now move on to discuss two null distribution estimation methods, namely the permutation approach and the Nyström spectral approach. The permutation approach is exactly the same as Sect. 2.4.1 with \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z}})\) replaced by \(\hat{\varXi }_{Ny, \tilde{k}_{\mathcal {X}},\tilde{k}_{\mathcal {Y}}}({\mathbf {z}})\). It is worth noting that for each permutation, we need to simulate a new set of inducing points for X and Y such that \(n_x, n_y \ll m\) with m being the number of samples.
Likewise, the Nyström spectral approach is similar to that described in Sect. 2.4.2 where eigendecompositions of the centred Gram matrices are required to simulate the null distribution. The difference is that we approximate the centred Gram matrices using Nyström method and the HSIC V-statistic is replaced by the Nyström HSIC estimator \(\hat{\varXi }_{Ny, \tilde{k}_{\mathcal {X}},\tilde{k}_{\mathcal {Y}}}({\mathbf {z}})\). So, the null distribution is then estimated using the eigenvalues from the covariance matrices \(\tilde{\varPhi }_X^T \tilde{\varPhi }_X \) and \( \tilde{\varPhi }_Y^T \tilde{\varPhi }_Y \). In such a way, the computational complexity is reduced from the original \({\mathcal {O}}(m^3)\) to \({\mathcal {O}}(n_x^3+n_y^3+(n_x^2+n_y^2)m + n_xn_ym)\) i.e. linear in m.
4.2 Random Fourier feature HSIC
So far, we have looked at two large-scale approximation techniques that are applicable to any positive-definite kernel. If the corresponding kernel also happens to be translation invariant with the moment condition in (39); however, an additional popular large-scale technique can be applied: random Fourier features of Rahimi and Recht (2007) which is based on Bochner’s representation. Note that many other kernels though not translational invariant, e.g. arc-cosine kernel, are also universal and approximable by random features (Cho and Saul 2009). In this section, we will first review Bochner’s theorem and subsequently build up to how random Fourier features can be used to approximate large kernel matrices. Utilising it in the context of independence testing, we propose the RFF HSIC estimator and further consider two null distribution estimation approaches.
4.2.1 Bochner’s theorem
Through the projection of data into lower-dimensional randomised feature space, Rahimi and Recht (2007) proposed a method of converting the training and evaluation of any kernel machine into the corresponding operations of a linear machine. In particular, they showed using a randomised feature map \(z: {\mathcal {R}}^d \rightarrow {\mathcal {R}}^D\) we can obtain
where \(x,y \in {\mathcal {R}}^d\). More specifically, Rahimi and Recht (2007) demonstrate the construction of feature spaces that uniformly approximate shift-invariant kernels with \(D = O(d \epsilon ^{-2} \log \frac{1}{\epsilon ^2})\) where \(\epsilon \) is the accuracy of approximation. However, as we will see later certain moment conditions need to be satisfied.
Bochner’s theorem provides the key observation behind such approximation. This classical theorem (Theorem 6.6 in Wendland 2005) is useful in several contexts where one deals with translation-invariant kernels k, i.e. \(k(x,y) = \kappa (x-y)\). As well as constructing large-scale approximation to kernel methods (Rahimi and Recht 2007), it can also be used to determine whether a kernel is characteristic, i.e. if the Fourier transform of a kernel is supported everywhere then the kernel is characteristic (Sriperumbudur 2010).
Theorem 2
Bochner’s theorem (Wendland 2005) A continuous transition-invariant kernel k on \({\mathcal {R}}^d\) is positive definite if and only if \(k(\delta )\) is the Fourier transform of a non-negative measure.
For a properly scaled transition-invariant kernel k, the theorem guarantees that its Fourier transform \(\varGamma (w)\) is a non-negative measure on \({\mathcal {R}}^d\). Without loss of generality, \(\varGamma \) is a probability distribution. Since we would like to approximate real-valued kernel matrices, let us consider the approximation which uses only real-valued features. \(\kappa (x-y)\) can be written as:
provided that
Note, (37) follows because kernels are real valued and (38) uses the double-angle formula for cosine. The random features can be computed by first sampling \(\{w_j\}^D_{j=1} \overset{i.i.d.}{\sim }\varGamma \) and then for \(x_j \in {\mathcal {R}}^d\) with \(j \in \{1, \ldots , n \}\), setting \(z(x_j) = \sqrt{\frac{2}{D}}( \cos (w_1^Tx_j),\sin (w_1^Tx_j), \ldots , \cos (w_{\frac{D}{2}}^Tx_j), \sin (w_{\frac{D}{2}}^Tx_j))\) for \(j \in \{1,\ldots ,n \}\).
Here, we deal with explicit feature space and apply linear methods to approximate the Gram matrix through the covariance matrix \(Z(x)^TZ(x)\) of dimension \(D \times D\) where Z(x) is the matrix of random features. Essentially, (39) guarantees that the second moment of the Fourier transform of this translational invariant kernel k to be finite and hence ensure the uniform convergence of \(z(x)^Tz(y)\) to \(\kappa (x-y)\) (Rahimi and Recht 2007).
4.2.2 RFF HSIC estimator
The derivation of the biased RFF HSIC estimator follows in the same manner as Sect. 4.1.1. However, with the RFF approximations of the kernel matrices, (24) becomes:
where \(Z_x \in {\mathbb {R}}^{m \times D_x}\) and \(Z_y \in {\mathbb {R}}^{m \times D_y}\). Hence, when RFF estimators are substituted, the cross-covariance operator is simply a \(D_x \times D_y\) matrix. In the same way as the Nyström HSIC estimator, the HSIC statistic computed using RFF is a HSIC statistic computed using a different kernel, i.e. one that is induced by the random features. It is worth noting that the analysis of convergence of such estimator can possibly be done similarly to the analysis by Sutherland and Schneider (2015) for MMD. However, we will leave this for future work.
To use the RFF HSIC statistic in independence testing, the permutation approach and spectral approach in the previous section can be adopted for null distribution estimation with \(\hat{\varXi }_{Ny, \tilde{k}_{\mathcal {X}},\tilde{k}_{\mathcal {Y}}}({\mathbf {z}})\) replaced by \(\hat{\varXi }_{RFF, \tilde{k}_{\mathcal {X}},\tilde{k}_{\mathcal {Y}}}({\mathbf {z}})\). Just as the case with inducing points, the \(\{w_j\}_{j=1}^{D_.}\) should be sampled each time independently for X and Y when the RFF approximations \(Z_x\) and \(Z_y\) needed to be computed. As a remark, the number of inducing points and the number of \(w_j\)s plays a similar role in both methods which controls the trade-off between computational complexity and statistical power. In practice, as we will demonstrate in the next section, such number can be much smaller than the size of the dataset without compromising the performance.
5 Experiments
In this section, we present three synthetic data experiments to study the behaviour of our large-scale HSIC tests. The main experiment is on a challenging nonlinear low signal-to-noise ratio dependence dataset to assess the numerical performance amongst the large-scale HSIC tests. To investigate the performance of these test in a small scale, we further conduct linear and sine dependence experiments to compare with currently established methods for independence testing. Throughout this section, we set the significance level of the hypothesis test to be \(\alpha = 0.05\). Both Type I and Type II errors are calculated based on 100 trials. The 95% confidence intervals are computed based on normality assumption, i.e. \(\hat{\mu } \pm 1.96 \sqrt{\frac{\hat{\mu } (1- \hat{\mu })}{100}}\), where \(\hat{\mu }\) is the estimate for the statistical power. Unless otherwise stated, HSIC, RFF and Nyström approaches are all using Gaussian kernel with median heuristic.
5.1 Simple linear experiment
We begin with an investigation of the performance of our methods on a toy example with a small number of observations, in order to check the agreements between large-scale approximation methods we proposed and the exact methods where they are still feasible. Towards this aim, we consider a simple linear dependence experiment, but where the dependence between the response Y and the input X is in only a single dimension of X. In particular,
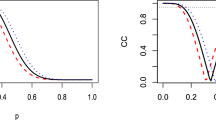
where d is the dimensionality of data vector X and \(X_1\) indicate the first dimension of X. The noise Z is independent standard Gaussian noise. We would like to compare methods based on HSIC to a method based on Pearson’s correlation which is explicitly aimed at linear dependence and should give strongest performance. However, as the latter cannot be directly applied to multivariate data, we consider a SubCorr statistic: SubCorr \(=\frac{1}{d}\sum ^d_{i=1} {\text {Corr}}(Y,X_i)^2\) where Corr(\(Y,X_i\)) is the Pearson’s correlation between Y and the \(i^{th}\) dimension of X . In addition, we will also consider SubHSIC statistic: SubHSIC\(=\frac{1}{d}\sum ^d_{i=1} {\text {HSIC}}(Y,X_i)^2\). For these two methods, we will use a permutation approach as their distributions are not immediately clear.
In Fig. 1, the dimension of X is set to be 10. Both the number of random features in RFF and the number of inducing variables in Nyström are set to 10. We do not use the block-based method as the sample sizes are small. From Fig. 1 (right), we see that SubCorr yields the highest power as expected. HSIC and SubHSIC with Gaussian median heuristic kernels perform similarly though, with all three giving the power of 1 at the sample size of 100. On the other hand, Fig. 1 (left) shows that the two large-scale methods are still able to detect the dependence at these small sample sizes, even though there is some loss in power in comparison to HSIC and they would require a larger sample size. As we will see, this requirement for a larger sample size will be offset by a much lower computational cost in large-scale examples.
5.2 Nonlinear experiments
In this section, we consider two nonlinear experiments: one with relatively small sample sizes (up to 4000 samples) and a large-scale scenario (with number of samples from 1000 to \(10^7\)). For both experiments, we investigate the power versus time trade-off of the introduced large-scale approximate tests and compare their performance to quadratic time ones. A summary of the methods investigated here is illustrated in Table 1.
In addition to HSIC and its large-scale versions, we will also consider a normalisation of HSIC: dCor (Székely et al. 2007; Székely and Rizzo 2009) which can be formulated in terms of HSIC using a Brownian Kernel with parameter \(h=0.5\) (Sejdinovic et al. 2013b, Appendix A). For clarity and consistency of comparison, methods using the same kernel will be compared with each other. In particular, we will compare HSIC and its large-scale approximations with GdCor (dCor using Gaussian kernel with median heuristic bandwidth parameter) and its corresponding large-scale approximations. As the asymptotic null distribution for GdCor is unclear, a permutation approach (see Sect. 2.4.1) will be used for p value computations. Similar comparison will be done for the Brownian kernel with \(h=0.5\).
Note that random Fourier feature approaches cannot be directly applied to the Brownian kernel as it is not translational invariant. In addition, we will not use the block-based approach in the small-scale nonlinear experiment as the asymptotic normality assumption for the null distribution requires large sample size—i.e. large number of blocks as well as a large number of samples per block.
5.2.1 Small-scale nonlinear experiment
Consider a sine dependence experiment to investigate time versus power trade-offs of large-scale tests. The dataset consists of a sample of size m generated i.i.d. according to:
where d is the dimensionality of data vector X, \(X_i\) indicates the \(i^{{\text {th}}}\) dimension of X and \(Z\sim {\mathcal {N}}(0,1)\). For this experiment, the dependency can be detected with a relatively small number of samples. Figure 2 illustrates the power as a function of the sample size when \(d=2\); the number of random Fourier features and inducing variables are here both set to 50.
From Fig. 2, dCor clearly outperforms all the other methods with its Nyström approximation giving the closest performance in terms of power. Reassuringly, all the six methods using Gaussian kernel give very similar power performance. Although dCor and its large-scale approximation give superior power performance, the performance of HSIC and its large-scale approximations seems to be indifferent to the kernel used. Figure 3, however, tells a much more interesting story—the large-scale methods all reach the power of 1 in a test time which is several orders of magnitude smaller, demonstrating the utility of the introduced tests.
5.2.2 Large-scale nonlinear experiment
Now, we would like to compare the performance of the proposed large-scale HSIC tests with each other—at sample sizes where standard HSIC/dCor approaches are no longer feasible. Gaussian kernel with median heuristic bandwidth will be used throughout this section contrasting the block-based spectral approach, RFF spectral approach and the Nyström spectral approach. The other approximate methods listed in Table 1 require costly permutation approaches to compute the null distribution, and we therefore do not examine them in this subsection.
We consider a challenging nonlinear and low signal-to-noise ratio experiment, where a sample of size m is generated i.i.d. according to:
where d is the dimensionality of the data set X and \(Z \sim {\mathcal {N}}(0,I_{\frac{d}{2}+1})\). Note that Y is independent of each individual dimension of X and that the dependence is nonlinear. For \(d = 50\) and 100, we would like to explore the relationship between the test power across a different number of samples \(m = \{ 10^5, 2\times 10^5, 5\times 10^5, 10^6, 2\times 10^6,5\times 10^6, 10^7 \}\). The number of random features, inducing variables and block size are all set to 200 so that their computational cost is comparable. Gaussian RBF kernel with median heuristic is used in all cases. For RFF and Nyström methods, we used the spectral approach to estimate the null distribution.
Large-Scale Experiment: The statistical power comparison between the three large-scale independence testing methods based on 100 trials. Dotted line \(d = 50\); solid line \(d=100\). The 95% confidence intervals are computed based on normality assumption, i.e. \(\hat{\mu } \pm 1.96 \sqrt{\frac{\hat{\mu } (1- \hat{\mu })}{100}}\), where \(\hat{\mu }\) is the estimate for the statistical power
Large-Scale Experiment: The average testing time comparison between the three large-scale independence testing methods for samples size \(m = \{ 10^5, 2\times 10^5, 5\times 10^5, 10^6, 2\times 10^6,5\times 10^6, 10^7 \}\). Quadratic time HSIC spectral method is also used on subset of data of size \(\{100, 200, 500, 1000, 2000, 4000\}\) as baseline comparison for similar testing time. Dotted line \(d = 50\); solid line \(d=100\)
Figure 4 is a plot of the test power against the number of samples whereas Fig. 5 is a plot of the test power against average testing time. In the computational time comparison plot, we also present baseline experiments which simply apply the quadratic time HSIC spectral approach on a subset of the available data (with a subset size be in \(\{100, 200, 500, 1000, 2000, 4000\}\)). For the considered computation budgets, it is clear that this approach is unable to detect the dependence.
It is clear in Fig. 4 that for both \(d=50\) and \( d = 100\), the RFF method gives the best performance in power for a fixed number of samples, followed by the Nyström method and then by the block-based approach. Although parallel computing could be employed for the block-based HSIC method, as we observed, RFF and Nyström methods will be preferred for achieving higher statistical power at any given samples size. The RFF method is able to achieve zero type II error (i.e. no failure to reject a false null) with 5\(\times 10^4\) samples for \(d=50\) and 5\(\times 10^5\) samples for \(d=100\), while the Nyström method has a 80% false negative rate at these sample sizes. The power versus time plot in Fig. 5 gives a similar picture as Fig. 4 confirming the superiority of the RFF method on this example.
5.2.3 Real data experiment
In this section, we examine the performance of the three proposed large-scale HSIC tests on real data—a subset of the Million Song Dataset (MSD) (Bertin-Mahieux et al. 2011). The dataset consists of 515,345 observations with each song X represented by 90 features, of which 12 features are timbre average (over all segments) of the song, and 78 features are timbre covariance. The aim is to detect the dependence between X and the year of release Y which ranges from 1922 to 2011. As such dependency is rather easy to detect (Jitkrittum et al. 2016), we jitter each entry of the X matrix with independent Gaussian noise with mean 0 and variance 1000. The significance level is set at \(\alpha =0.05\) and we repeat for 200 trials where each time a subset of the data is randomly sampled. The number of random Fourier features, inducing points and block size are all set to be 10. Figure 6 shows the power performance across a range of sample sizes. For this dataset, Nyström spectral approach provides the best power performance followed by RFF spectral approach. Similar to the previous experiment, the Block-based method gives the least power for any fixed number of samples. The power versus average testing time plot gives a similar picture.
Real Data Experiment: (left) power versus sample size; (right) power versus average testing time. The statistical power comparison between the three large-scale independence testing methods based on 200 trials. The 95% confidence intervals are computed based on normality assumption, i.e. \(\hat{\mu } \pm 1.96 \sqrt{\frac{\hat{\mu } (1- \hat{\mu })}{100}}\), where \(\hat{\mu }\) is the estimate for the statistical power
6 Discussion and conclusions
We have proposed three large-scale estimators of HSIC, a kernel-based nonparametric dependence measure—these are the block-based estimator, the Nyström estimator and the RFF estimator. We subsequently established suitable independence testing procedures for each method—by taking advantage of the normal asymptotic null distribution of the block-based estimator and by employing an approach that directly estimates the eigenvalues appearing in the asymptotic null distribution for the Nyström and RFF methods. All three tests significantly reduce computational complexity in memory and time over the standard HSIC-based test. We verified the validity of our large-scale testing methods and its favourable trade-offs between testing power and computational complexity on challenging high-dimensional synthetic data. We have observed that RFF and Nyström approaches have considerable advantages over the block-based test. Several further extensions can be studied: the developed large-scale approximations are readily applicable to three-variable interaction testing (Sejdinovic et al. 2013a), conditional independence testing (Fukumizu et al. 2008) as well as application in causal discovery (Zhang et al. 2011; Flaxman et al. 2015). Moreover, the RFF HSIC approach can be extended using the additional smoothing of characteristic function representations similarly to the approach of Chwialkowski et al. (2015) in the context of two-sample testing. Furthermore, one can also consider the robustness of the proposed approaches in the case where heterogeneous datasets are considered, or when some form of within-sample dependence is introduced, e.g. when testing for independence between time series (Chwialkowski and Gretton 2014).
Notes
An alternative approach applicable to scalar variables uses kernel-based methods combined with a copula transformation (Póczos et al. 2012) can be used to tabulate the null distribution, but this approach is not straightforward to generalise to modelling dependence between random vectors.
A straightforward estimator of dCor (Székely et al. 2007; Székely and Rizzo 2009) is then given by normalising \(\varXi _b({\mathbf {z}})\) by the Frobenius norms of \(HK_xH\) and \(HK_yH\), i.e. \(\widehat{dCor}(\mathbf{z}) = \frac{\langle HK_xH, HK_yH \rangle }{\Vert HK_xH\Vert _F\Vert HK_yH\Vert _F}\).
For example, \(B=m^\delta \ {\text {with }} \delta \in (0,1)\).
References
Anderson, N.H., Hall, P., Titterington, D.M.: Two-sample test statistics for measuring discrepancies between two multivariate probability density functions using kernel-based density estimates. J. Multivar. Anal. 50, 41–54 (1994)
Arcones, M.A., Gine, E.: On the bootstrap of \(U\) and \(V\) statistics. Ann. Stat. 20(2), 655–674 (1992)
Aronszajn, N.: Theory of reproducing kernels. Trans. Am. Math. Soc. 68(3), 337–404 (1950)
Bach, F., Jordan, M.I.: Kernel independent component analysis. J. Mach.Learn. 10, 1–48 (2002)
Berlinet, A., Thomas-Agnan, C.: Reproducing Kernel Hilbert Spaces in Probability and Statistics. Kluwer, Boston (2004)
Bertin-Mahieux, T., Ellis, D.P., Whitman, B., Lamere., P.: The million song dataset. In: International Conference on Music Information Retrieval (ISMIR) (2011)
Blaschko, M., Gretton, A.: Learning taxonomies by dependence maximization. Adv. Neural Inf. Process. Syst. 21, 153–160 (2009)
Borgwardt, K.M., Gretton, A., Rasch, M.J., Kriegel, H.P., Schlkopf, B., Smola, A.J.: Integrating structured biological data by kernel maximum mean discrepancy. Bioinformatics 22(14), e49–e57 (2006). doi:10.1093/bioinformatics/btl242
Cho, Y., Saul, L.K.: Kernel methods for deep learning. In: Bengio, Y., Schuurmans, D., Lafferty, J.D., Williams, C.K.I., Culotta, A. (eds.) Advances in Neural Information Processing Systems, vol. 22, pp. 342–350. Curran Associates Inc., Red Hook (2009)
Chwialkowski, K., Gretton, A.: A kernel independence test for random processes. In: Proceedings of the 31st International Conference on Machine Learning (2014)
Chwialkowski, K., Ramdas, A., Sejdinovic, D., Gretton, A.: Fast two-sample testing with analytic representations of probability measures. In: Advances in Neural Information Processing Systems (NIPS), vol. 28 (2015)
Cortes, C., Mohri, M., Rostamizadeh, A.: Algorithms for learning kernels based on centered alignment. J. Mach. Learn. Res. 13(1), 795–828 (2012)
Dauxois, J., Nkiet, G.M.: Nonlinear canonical analysis and independence tests. Ann. Stat. 26(4), 1254–1278 (1998)
Flaxman, S.R., Neill, D.B., Smola, A.J.: Gaussian processes for independence tests with non-iid data in causal inference. ACM Trans. Intell. Syst. Technol. 7(2), 22:1–22:23 (2015)
Fukumizu, K., Gretton, A., Sun, X., Schölkopf, B.: Kernel measures of conditional dependence. In: In Adv. NIPS (2008)
Gretton, A., Bousquet, O., Smola, A., Schölkopf, B.: Measuring Statistical Dependence with Hilbert–Schmidt Norms. Lecture Notes in Computer Science, pp. 63–77 (2005)
Gretton, A., Fukumizu, K., Schölkopf, B., Teo, C.H., Song, L., Smola, A.J.: A kernel statistical test of independence. In: NIPS (2008)
Gretton, A., Fukumizu, K., Harchaoui, Z., Sriperumbudur, B.: A fast, consistent kernel two-sample test. In: Advances in Neural Information Processing Systems, vol. 22. Curran Associates Inc., Red Hook, pp. 673–681 (2009). papers/1006_paperlong.pdf
Gretton, A., Sriperumbudur, B., Sejdinovic, D., Strathmann, H., Balakrishman, S., Pontil, M., Fukumizu, K.: Optimal kernel choice for large-scale two-sample tests. In: Advances in Neural Information Processing Systems (2012a)
Gretton, A., Borgwardt, K.M., Rasch, M.J., Schölkopf, B., Smola, A.: A kernel two-sample test. J. Mach. Learn. Res. 13, 723–773 (2012b)
Huang, S.Y., Lee, M.H., Hsiao, C.K.: Nonlinear measures of association with kernel canonical correlation analysis and applications. J. Stat. Plan. Inference 139(7), 2162–2174 (2009)
Jitkrittum, W., Szabo, Z., Gretton, A.: An Adaptive Test of Independence with Analytic Kernel Embeddings (2016)
Lai, P., Fyfe, C.: Kernel and nonlinear canonical correlation analysis. Int. J. Neural Syst. 10(5), 365–377 (2000)
Lopez-Paz, D.: From Dependence to Causation. Ph.D. thesis, University of Cambridge (2016)
Lopez-Paz, D., Hennig, P., Schölkopf, B.: The randomized dependence coefficient. Adv. Neural Inf. Process. Syst. 26, 1–9 (2013)
Lopez-Paz, D., Sra, S., Smola, A., Ghahramani, Z., Schölkopf, B.: Randomized nonlinear component analysis. In: Proceedings of the 31st International Conference on Machine Learning, pp. 1359–1367 (2014)
Lyons, R.: Distance covariance in metric spaces. Ann. Probab. 41(5), 3284–3305 (2013)
Mardia, K., Kent, J., Bibby, J.: Multivariate Analysis. Academic Press, New York (1979)
Nguyen, D., Eisenstein, J.: A Kernel Independence Test for Geographical Language Variation (2016). ArXiv:1601.06579
Peters, J., Mooij, J.M., Janzing, D., Schölkopf, B.: Causal discovery with continuous additive noise models. J. Mach. Learn. Res. 15(1), 2009–2053 (2014)
Póczos, B., Ghahramani, Z., Schneider, J.G.: Copula-based kernel dependency measures. In: Proceedings of the 29th International Conference on Machine Learning, ICML 2012, Edinburgh, Scotland, UK, June 26–July 1, 2012 (2012)
Rahimi, A., Recht, B.: Random Features for Large-Scale Kernel Machines. Adv. Neural Inf. Process. Syst. 20 (2007)
Reed, M., Simon, B.: Methods of Modern Mathematical Physics. I: Functional Analysis, 2nd edn. Academic Press, New York (1980)
Rubenstein, P.K., Chwialkowski, K.P., Gretton, A.: A kernel test for three-variable interactions with random processes. arXiv preprint (2015). ArXiv:1603.00929
Schölkopf, B., Smola, A.: Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press, Cambridge (2002)
Sejdinovic, D., Gretton, A., Bergsma, W.: A kernel test for three-variable interactions. Adv. Neural Inf. Process. Syst. (NIPS) 26, 1124–1132 (2013a)
Sejdinovic, D., Sriperumbudur, B., Gretton, A., Fukumizu, K.: Equivalence of distance-based and RKHS-based statistics in hypothesis testing. Ann. Stat. 41(5), 2263–2291 (2013b)
Sejdinovic, D., Strathmann, H., De, S., Zaremba, W., Blaschko, M., Gretton., A.: Big Hypothesis Tests with Kernel Embeddings: An Overview. Technical Report (2014). Gatsby Unit, UCL
Serfling, R.J.: Approximation Theorems of Mathematical Statistics. Wiley, New York (2002)
Smola, A., Gretton, A., Song, L., Schölkop, B.: A Hilbert space embedding for distributions. In: Algorithmic Learning Theory: 18th International Conference, pp. 13–31 (2007)
Snelson, E., Ghahramani, Z.: Sparse Gaussian processes using pseudo-inputs. In: Advances in Neural Information Processing Systems, vol. 18, pp. 1257–1264. MIT press (2006)
Song, L., Smola, A., Gretton, A., Borgwardt, K.M.: A dependence maximization view of clustering. In: Proceedings of the 24th International Conference on Machine Learning, pp. 815–822 (2007)
Song, L., Smola, A., Gretton, A., Bedo, J., Borgwardt, K.: Feature selection via dependence maximization. J. Mach. Learn. Res. 13, 1393–1434 (2012). http://jmlr.csail.mit.edu/papers/v13/song12a.html
Sriperumbudur, B.K.: Reproducing Kernel Space Embeddings and Metrics on Probability Measures. PhD Thesis, University of California, San Diego (2010)
Steinwart, I., Christmann, A.: Support Vector Machines. Springer, New York (2008)
Sutherland, D.J., Schneider, J.: On the error of random fourier features. In: Conference on Uncertainty in Artificial Intelligence (UAI) (2015)
Székely, G.J., Rizzo, M.L.: Brownian distance covariance. Ann. Appl. Stat. 3(4), 1236–1265 (2009)
Székely, G.J., Rizzo, M.L., Bakirov, N.K.: Measuring and testing dependence by correlation of distances. Ann. Stat. 35(6), 2769–2794 (2007)
Wendland, H.: Scattered Data Approximation. Cambridge University Press, Cambridge (2005)
Williams, C.K.I., Seeger, M.: Using the Nyström method to speed up kernel machines. In: Leen, T., Dietterich, T., Tresp, V. (eds.) Advances in Neural Information Processing Systems, vol. 13, pp. 682–688. MIT Press (2001). http://papers.nips.cc/paper/1866-using-the-nystrom-method-to-speed-up-kernel-machines
Zaremba, A., Aste, T.: Measures of causality in complex datasets with application to financial data. Entropy 16(4), 2309 (2014)
Zaremba, W., Gretton, A., Blaschko, M.: B-test: a non-parametric, low variance kernel two-sample test. In: Advances in Neural Information Processing Systems (2013)
Zhang, K., Peters, J., Janzing, D., Schölkopf, B.: Kernel-based conditional independence test and application in causal discovery. In: Uncertainty in Artificial Intelligence, pp. 804–813 (2011)
Zhao, J., Meng, D.: FastMMD: ensemble of circular discrepancy for efficient two-sample test. Neural Comput. 27(6), 1345–1372 (2015)
Author information
Authors and Affiliations
Corresponding author
Additional information
QZ was supported by the EPSRC (EP/M50659X/1). DS was supported in part by the ERC (FP7/617071).
Appendix: Proof of Theorem 1
Appendix: Proof of Theorem 1
We note that Lyons (2013) gives a proof to a similar theorem (Theorem 2.7 in Lyons 2013) regarding generalised versions of distance covariance (dCov). We closely follow the steps given in Proposition 2.6 and Theorem 2.7 of Lyons (2013). However, the proof provided here is tailored to the kernel view of HSIC/dCov duality (Sejdinovic et al. 2013b) and it is slightly more general in that it applies to any semimetric (rather than metric) of negative type. Unless stated otherwise, the notation follows that used in the main part of this paper.
Recall from the main part of this paper that the existence of the HSIC statistics \(\varXi _{k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {z}})\) defined in (6) requires the marginal distributions to have finite first moment with respect to the kernels, i.e. \(P_X \in {\mathcal {M}}^1_{k_{{\mathcal {X}}}}({\mathcal {X}})\) and \(P_Y \in {\mathcal {M}}^1_{k_{{\mathcal {Y}}}}({\mathcal {Y}})\). By Proposition 20 of Sejdinovic et al. (2013b), this translates directly into finite first moment conditions with respect to the semimetrics: \(P_X \in {\mathcal {M}}^1_{d_x}({\mathcal {X}})\) and \(P_Y \in {\mathcal {M}}^1_{d_y}({\mathcal {Y}})\) when \(k_{{\mathcal {X}}}\) generates \(d_x\) and \(k_{{\mathcal {Y}}}\) generates \(d_y\).
More specifically, a valid semimetric d of negative type on \({\mathcal {Z}}\) generated by a non-degenerate kernel k on \({\mathcal {Z}}\) can be written as \(d(z,z') = k(z,z) + k(z',z')-2k(z,z')\) (Corollary 16 Sejdinovic et al. 2013b). Then, such semimetric \(d_x\) centred at the probability measure \(P_X\) defined on \({\mathcal {X}}\) is
Similarly, \(d_{P_Y} (y, y')\) is the semimetric centred at the probability measure \(P_Y\) defined on \({\mathcal {Y}}\). If we substitute the kernel representation of the semimetric for X and Y, respectively, into \(d_{P_X} (x,x')\) and \(d_{P_Y} (y,y')\), we obtain the following:
where \(\tilde{k}_{P_X}(x,x')\) and \( \tilde{k}_{P_Y}(y,y') \) are defined in (12).
Let \((X^i,Y^i) \sim \theta \) be independent for \( i \in \{1,2,\ldots ,6\}\), we introduce the “core” defined in Lyons (2013),
where for \(z_i \in {\mathcal {X}}\) or \(z_i \in {\mathcal {Y}}\):
The second line follows from the relationship between d and k where \(k_{P_X}\) is used for \(z_i \in {\mathcal {X}}\) and \(k_{P_Y}\) for \(z_i \in {\mathcal {Y}}\).
In fact, we can prove that the expectation of (44) is four times the HSIC of X and Y. To see this, we first need to show that such expectation is well defined. Indeed, note that for a valid semimetric d of negative type on \({\mathcal {Z}}\), \(\sqrt{d(z,z')} = ||k(\cdot ,z) - k(.,z') ||_{{\mathcal {H}}_k}\), then the following inequality holds:
It then follows that
and that
Hence, replacing the \(z_i\) in \(g_1\) with \(X^i\) and the \(z_i\) in \(g_2\) with \(Y^i\),
Since the marginal distributions have finite first moments with respect to the kernels, then each of the terms in \(g_1\) and \(g_2\) is integrable and hence \(g_1\) and \(g_2\) are integrable. Moreover, since the marginal distributions have finite second moments with respect to the kernels, then the joint distribution satisfies \(P_{XY} \in {\mathcal {M}}^1_{k_{{\mathcal {X}}} \otimes k_{{\mathcal {Y}}}}({\mathcal {X}}\times {\mathcal {Y}})\). Therefore, h is integrable. Subsequently, by taking the expectation and utilising Fubini’s theorem, we obtain that
which is 4 times the HSIC.
In order to use the theory of degenerate V-statistics to obtain the asymptotic distribution, we need to consider the symmetrised version of h, which we define as follows
where A denotes the set of all permutations of \(\{1,\ldots ,6\}\). Then, under the null hypothesis of independence \(P_{XY} = P_X \times P_Y \),
If we fix the first two positions to be \(\{1,2\}\) and randomly permute the rest, we obtain 24 different combinations. Similarly if we fix the first two positions to be \(\{2,1\}\), we also obtain 24 different combinations. Some algebraic manipulation shows that these are the combinations that gives the expectation of h to be \(\tilde{k}_\mu (x,x') \tilde{k}_\nu (y,y').\) In fact, these are the only combinations as when either \(\{1\}\) or \(\{2\}\) or both are not in the first two positions, the expectation of h is zero and all terms cancel out.
Another important condition to check is that \(\bar{h}_2\) has finite second moment. It was shown earlier that \(h((X^1,Y^1), \ldots , (X^6,Y^6))\) of (44) is integrable. Subsequently, \(f(X^1,X^2,X^3,X^4)\) has finite second moment. Hence, \(h((X^1,Y^1), \ldots , (X^6,Y^6))\) has finite second moment under the null hypothesis. Additionally, by Jensen’s inequality, \({\mathbb {E}}(|\bar{h}_2 ((X,Y),(X,Y))|) \le \{{\mathbb {E}}((\bar{h}_2 ((X,Y),(X,Y)))^2) \}^{1/2} < \infty \).
Hence, by Theorem B in Chap. 6 of Serfling (2002), which says
as the sample size \(m \rightarrow \infty \), we obtain that
with \(Z_r \overset{i.i.d.}{\sim }{\mathcal {N}}(0,1) \ \forall r\) and \(\{ \gamma _r \}^\infty _{r=1}\) are the eigenvalues of the operator \(S_{\tilde{k} }\): \(L^2_\theta ({\mathcal {X}} \times {\mathcal {Y}}) \rightarrow L^2_\theta ({\mathcal {X}} \times {\mathcal {Y}})\) defined as:
Under the null hypothesis \(P_{XY} = P_X \times P_Y\), the above operator is given by the tensor product of \(S_{\tilde{k}_{P_X}}\) and \(S_{\tilde{k}_{P_Y}}\) (Remark 2.9 Lyons 2013). Therefore, \(\{ \gamma _r \}^\infty _{r=1}\) are the products of the eigenvalues of these two operators. Recall that \(k_{\mathcal {X}}\) is equivalent to \(\tilde{k}_{P_X}\) and \(k_{\mathcal {Y}}\) is equivalent to \(\tilde{k}_{P_Y}\) in the sense of Sejdinovic et al. (2013b). By noting that \(\varXi _{b,k_{\mathcal {X}},k_{\mathcal {Y}}}({\mathbf {Z}})\) is invariant under the change of kernel within the class of equivalent kernels, \(\frac{1}{m^2} \sum _{i,j} \tilde{k}_{P_X}(x_i,x_j) \tilde{k}_{P_Y} (y_i,y_j)\) is the highest order term in the V-statistics and thus determine its asymptotic null distribution.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhang, Q., Filippi, S., Gretton, A. et al. Large-scale kernel methods for independence testing. Stat Comput 28, 113–130 (2018). https://doi.org/10.1007/s11222-016-9721-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-016-9721-7