Abstract
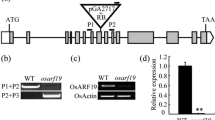
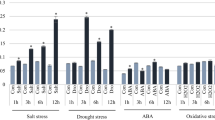
T-DNA-tagged rice plants were screened under cold- or salt-stress conditions to determine the genes involved in the molecular mechanism for their abiotic-stress response. Line 0-165-65 was identified as a salt-responsive line. The gene responsible for this GUS-positive phenotype was revealed by inverse PCR as OsGSK1 (O ryza s ativa g lycogen s ynthase k inase3-like gene 1), a member of the plant GSK3/SHAGGY-like protein kinase genes and an orthologue of the Arabidopsis b rassinosteroid in sensitive 2 (BIN2), AtSK21. Northern blot analysis showed that OsGSK1 was most highly detected in the developing panicles, suggesting that its expression is developmental stage specific. Knockout (KO) mutants of OsGSK1 showed enhanced tolerance to cold, heat, salt, and drought stresses when compared with non-transgenic segregants (NT). Overexpression of the full-length OsGSK1 led to a stunted growth phenotype similar to the one observed with the gain-of-function BIN/AtSK21 mutant. This suggests that OsGSK1 might be a functional rice orthologue that serves as a negative regulator of brassinosteroid (BR)-signaling. Therefore, we propose that stress-responsive OsGSK1 may have physiological roles in stress signal-transduction pathways and floral developmental processes.










Similar content being viewed by others
References
Alvarado MC, Zsigmond LM, Kovacs I, Cseplo A, Koncz C, Szabados LM (2004) Gene trapping with firefly luciferase in Arabidopsis: tagging of stress-responsive genes. Plant Physiol 134:18–27
Bray EA, Bailey-Serres J, Weretilnyk E (2000) Response to abiotic stresses. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists Press, Rockville, MD, pp 1158–1203
Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130:577–590
Chen DH, Roland PC (1999) A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep 17:53–57
Chen GP, Ma WS, Huang ZJ, Xu T, Xue YB, Shen YZ (2003) Isolation and characterization of TaGSK1 involved in wheat salt tolerance. Plant Sci 165:1369–1375
Cho HT, Kende H (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9:1661–1671
Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26:573–582
Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol 130:1506–1515
Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol 133:1209–1219
Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:4985–4990
Claes B, Dekeyser R, Villarroel R, van den Bulcke M, Bauw G, van Montagu M, Caplan A (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2:19–27
Clouse SD, Sasse JM (1998) Brassinosteroids: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49:427–451
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469
Dai Z, Gao J, An K, Lee JM, Edwards GE, An G (1996) Promoter element controlling developmental and environmental regulation of a tobacco ribosomal protein gene L34. Plant Mol Biol 32:1055–1065
Dornelas MC, Lejeune B, Dron M, Kreis M (1998) The Arabidopsis SHAGGY-related protein kinase (ASK) gene family: structure, organization and evolution. Gene 212:249–257
Dornelas MC, van Lammeren AA, Kreis M (2000) Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J 21:419–429
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke D (1991) Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3:149–157
Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990:87–92
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jeong SW, Choi SM, Lee DS, Ahn SN, Hur Y, Chow WS, Park YI (2002) Differential susceptibility of photosynthesis to light-chilling stress in rice (Oryza sativa L.) depends on the capacity for photochemical dissipation of light. Mol Cells 13:419–428
Jonak C, Beisteiner D, Beyerly J, Hirt H (2000) Wound-induced expression and activation of WIG, a novel glycogen synthase kinase 3. Plant Cell 12:1467–1475
Jonak C, Hirt H (2002) Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an merging family with novel functions. Trends Plant Sci 7:457–461
Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H (1996) Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA 93:11274–11279
Kagale S, Divi UK, Kronchko JE, Keller WA, Krishna P (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225:354–364
Katiyar-Agarwal S, Agarwal M, Grover A (2003) Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol 51:677–686
Kim ST, Kim SG, Hwang DH, Kang SY, Koo SC, Cho MJ, Kang KY (2004) Expression of a salt-induced protein (SALT) in suspension-cultured cells and leaves of rice following exposure to fungal elicitor and phytohormones. Plant Cell Rep 23:256–262
Krishna P (2003) Brassinosteroid-mediated stress responses. J Plant Growth Regul 22:289–297
Kwon M, Choe S (2005) Brassinosteroid biosynthesis and dwarf mutant. J Plant Biol 48:1–15
Lee SC, Kim JY, Kim SH, Lee K, Han SK, Choi HS, Jeong DH, An G (2003a) Trapping and characterization of cold-responsive genes from T-DNA tagging lines in rice. Plant Sci 166:69–79
Lee S, Kim J, Son JS, Nam J, Jeong DH, Lee K, Jang S, Yoo J, Lee J, Lee DY, Kang HG, An G (2003b) Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol 44:1403–1411
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:825–827
Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295:1299–1301
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Mandava NB (1988) Plant growth promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39:23–52
McElroy D, Rothernberg MM, Wu R (1990) Structural characterization of a rice actin gene. Plant Mol Biol 14:163–171
Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ (2002) Cloning the tomato Curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plants steroid hormone and peptide hormone signaling. Plant Cell 14:3163–3176
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) Gene Doc analysis and visualization of genetic variation. EMBNEW NEWS 4:14
Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T (2003) The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J 36:291–300
Orena SJ, Torchia AJ, Garofalo RS (2000) Inhibition of glycogen-synthase kinase 3 stimulates glycogen synthase and glucose transport by distinct mechanisms in 3T3-L1 adipocytes. J Biol Chem 275:15765–15772
Pay A, Jonak C, Bogre L, Meskiene I, Mairinger T, Szalay A, Heberle-Bors E, Hirt H (1993) The MsK family of alfalfa protein kinase genes encodes homologues of shaggy/glycogen synthase kinase-3 and shows differential expression patterns in plant organs and development. Plant J 3:847–856
Perez-Perez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242:161–173
Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I (2001) Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J 27:305–314
Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I (1999) An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol 119:1527–1534
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and cold: differences and cross-talk between two stress signal pathways. Curr Opin Plant Biol 3:217–223
Sun Y, Allen RD (2005) Functional analysis of the BIN 2 genes of cotton. Mol Genet Genom 274:51–59
Sun Y, Fokar M, Asami T, Yoshida S, Allen RD (2004) Characterization of the Brassinosteroid insensitive 1 genes of cotton. Plant Mol Biol 54:221–232
Takakura Y, Ito T, Saito H, Inoue T, Komari T, Kuwata S (2000) Flower-predominant expression of a gene encoding a novel class I chitinase in rice (Oryza sativa L.). Plant Mol Biol 42:883–897
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Tichtinsky G, Tavares R, Takvorian A, Schwebel-Dugue N, Twell D, Kreis M (1998) An evolutionary conserved group of plant GSK-3/shaggy-like protein kinase genes preferentially expressed in developing pollen. Biochim Biophys Acta 1442:261–273
Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M (2000) Characterization of XET-related genes of rice. Plant Physiol 122:853–859
Vert G, Chory J (2006) Downstream nuclear events in brassinosteroid signaling. Nature 441:96–100
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1606
Yang G, Komatsu S (2004) Microarray and proteomic analysis of brassinosteroid and gibberellin-regulated gene and protein expression in rice. Genom Prot Bioinfo 2:77–83
Yang G, Matsuoka M, Iwasaki Y, Komatsu S (2003) A novel brassinolide enhanced gene identified by cDNA microarray is involved in the growth of rice. Plant Mol Biol 52:843–854
Yoo MJ, Albert VA, Soltis PS, Soltis DE (2006) Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY-like kinase genes in plants. BMC Plant Biol 6:3
Yoshida S, Foro DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. IRRI, Los Baños, Philippines
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G., Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G., Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Zumbrunn J, Kinoshita K, Hyman AA, Nathke IS (2001) Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr Biol 11:44–49
Acknowledgments
We thank Priscilla Licht for critical proofreading of the manuscript. This work was funded in part by grants from the Biogreen 21 Program, Rural Development Administration and from the National Research Laboratory Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Serry Koh and Sang-Choon Lee are co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2007_9213_MOESM1_ESM.tif
Northern blot analysis of SalT under BL treatment. SalT expression level was increased to maximum at 24 h after BL treatment, suggesting SalT is a BL-responsive gene. C, Control; BL, Brassinolide (TIF 550 kb)
Rights and permissions
About this article
Cite this article
Koh, S., Lee, SC., Kim, MK. et al. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol 65, 453–466 (2007). https://doi.org/10.1007/s11103-007-9213-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9213-4