Abstract
Plant natural products (PNP) (e.g., secondary vegetal metabolites and their derivatives) have been a productive source of active ingredients for the pharmaceutical industry. The High Throughput Screening of Plant Natural Products (PNP-HTS) with extracts or isolated compounds has shown to be time consuming, expensive, and not as successful as expected. Recently building upon the innovative fragment-based drug discovery (FBDD) a disruptive approach was developed based on PNP. The fragment approach involves elaboration and/or isolation of weakly binding small molecules with molecular weights between 150 and 250 Da. This method is fundamentally different from HTS in almost every aspect (i.e., size of the compound library, screening methods, and optimization steps from hit to lead). Due to their nature, vegetal natural fragments have unique three-dimensional (3D) properties, high Fsp3, low aromaticity, and large chemo-diversities which represent potential opportunities for developing novel drugs. Preliminary results using vegetal natural fragments appear to be a promising and emerging field which offers valuable prospects for developing new drugs.


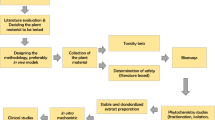
From David and Ausseil (2014)





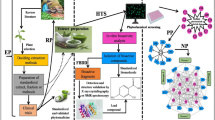
From Valenti et al. (2019)






Similar content being viewed by others
Abbreviations
- ADMET:
-
Administration–distribution–metabolization–excretion–toxicology studies
- FBDD:
-
Fragment based drug discovery
- Fsp3 :
-
Ratio of sp3 carbon to the total number of carbon
- HTS:
-
High throughput screening
- MS:
-
Mass spectrometry
- NMR:
-
Nuclear magnetic resonance
- NP:
-
Natural products
- PF:
-
Pierre Fabre
- PFL:
-
Plant fragment library
- PNP:
-
Plant natural products
- SPR:
-
Surface plasmon resonance
References
Agarwal G, Carcache PJB, Addo EM et al (2019) Current status and contemporary approaches to the discovery of antitumor agents from higher plants. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2019.01.004
Amirkia V, Heinrich M (2015) Natural products and drug discovery: a survey of stakeholders in industry and academia. Front Pharmacol 6:1–8
Atanasov AG, Waltenberger B, Pferschy-Wenzig EM et al (2015) Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv 33:1582–1614
Baell J, Walters MA (2014) Chemical con artists foil drug discovery. Nature 513:481–483
Baker M (2013) Fragment-based lead discovery grows up. Nat Rev Drug Discov 12:5–7
Baker M (2017) Fragment-based phenotypic screening is a hit. Nat Rev Drug Discov 16:225–226
Barelier S, Pons J, Gehring K, Lancelin JM et al (2010) Ligand specificity in fragment-based drug design. J Med Chem 53:5256–5266
Bathula SR, Akondi SM, Mainkar PS et al (2015) “Pruning of biomolecules and natural products (PBNP)”: an innovative paradigm in drug discovery. Org Biomol Chem 13:6432–6448
Blundell TL (2017) Protein crystallography and drug discovery: recollections of knowledge exchange between academia and industry. IUCrJ 4:308–321
Boufridi A, Quinn RJ (2017) Harnessing the properties of natural products. Annu Rev Pharmacol Toxicol 58:451–470
Breinbauer R, Vetter IR, Waldmann H (2002) From protein domains to drug candidates: natural products as guiding principles in the design and synthesis of compound libraries. Angew Chem Int Ed 41:2878–2890
Brenner S, Lerner RA (1992) Encoded combinatorial chemistry. Proc Natl Acad Sci USA 89:5381–5383
Brown DG, Boström J (2018) Where do recent small molecule clinical development candidates come from? J Med Chem 61:9442–9468
Butler MS, Fontaine F, Cooper MA (2014) Natural product libraries: assembly, maintenance, and screening. Planta Med 80:1161–1170
Carletti I, Massiot G, Debitus C (2011) Polyketide molecules as anticancer agents. PCT patent WO2011051380
Chan DSH, Whitehouse AJ, Coyne AG et al (2017) Mass spectrometry for fragment screening. Essays Biochem 61:465–473
Chanana S, Thomas CS, Braun DR et al (2017) Natural product discovery using planes of principal component analysis in R (PoPCAR). Metabolites 7(E34):1–12
Chavanieu A, Pugnière M (2016) Developments in SPR fragment screening. Expert Opin Drug Discov 11:489–499
Chen Y, de Bruyns Kops C, Kirchmair J (2017) Data resources for the computer-guided discovery of bioactive natural products. J Chem Inf Model 57:2099–2111
Chen Y, Gardia de Lomana M, Friedrich NO et al (2018) Characterization of the chemical space of known and readily obtainable natural products. J Chem Inf Model 58:1518–1532
Cherry M, Mitchell T (2008) Introduction to fragment-based drug discovery. In: Zartler ER, Shapiro MJ (eds) Chapter 1 in fragment-based drug discovery: a practical approach. Wiley, New York
Chessari G, Woodhead AJ (2009) From fragment to clinical candidate: a historical perspective. Drug Discov Today 14:668–675
Congreve M, Carr R, Murray C et al (2003) A ‘rule of three’ for fragment-based lead discovery. Drug Discov Today 8:876–877
Congreve M, Chessari G, Tisi D et al (2008) Recent developments in fragment-based discovery. J Med Chem 51:3661–3680
Crane EA, Gademann K (2016) Capturing biological activity in natural product fragments by chemical synthesis. Angew Chem Int Ed Engl 55:3882–3902
Dalvit C, Vulpetti A (2018) Ligand-based fluorine NMR screening: principles and applications in drug discovery projects. J Med Chem. https://doi.org/10.1021/acs.jmedchem.8b0121(Just Accepted Manuscript Publication)
Dalvit C, Pevarello P, Tatò M, Veronesi M, Vulpetti A, Sundström M (2000) Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biomol NMR 18:65–68
David B (2018) New regulations for accessing plant biodiversity samples, what is ABS? Phytochem Rev 17:1211–1223
David B, Ausseil F (2014) High-throughput screening of vegetal natural substances. In: Hostettmann K, Chen S, Marston A, Stuppner H (eds) Handbook of chemical and biological plant analytical methods. Wiley, New York, pp 987–1010 (Chapter 44)
David B, Wolfender JL, Dias DA (2015) The pharmaceutical industry and natural products: historical status and new trends. Phytochem Rev 14:299–315
Davis AM, Plowright AT, Valeur E (2017) Directing evolution: the next revolution in drug discovery? Nat Rev Drug Discov 16:681–698
Deyon-Jung L, Morice C, Chéry F et al (2016) Fragment pharmacophore-based in silico screening: a powerful approach for efficient lead discovery. Med Chem Commun 7:506–511
DiMasi JA, Grabowski HG, Hansen RW (2016) Innovation in the pharmaceutical industry: new estimates of R&D cost. J Health Econ 47:20–33
DNP (2017) Dictionary of natural products on CD-ROM. Chapman and Hall, Boca Raton, FL
Erlanson DA, Jahnke W (2006) The concept of fragment-based drug discovery (chapter 1). In: Jahnke W, Erlanson DA, Mannhold R, Kubinyi H, Folkers G (eds) Fragment-based approaches in drug discovery. Methods and principles in medicinal chemistry. Wiley, New York, pp 3–10
Erlanson DA, Zartler (2019) Practical fragments. http://practicalfragments.blogspot.com/. Accessed 28 Jan 2019
Erlanson DA, Fesik SW, Hubbard RE et al (2016) Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov 15:605–619
Fattori D (2004) Molecular recognition: the fragment approach in lead generation. Drug Discov Today 9:229–238
Feher M, Schmidt JM (2003) Properties distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci 43:218–227
Fischer M, Hubbard RE (2009) Fragment-based ligand discovery. Mol Interv 9:22–30
Fuller N, Spadola L, Cowen S et al (2016) An improved model for fragment-based lead generation at AstraZeneca. Drug Discov Today 21:1272–1283
Galloway WR, Spring DR (2011) Better leads come from diversity. Nature 470:43
Galloway WR, Isidro-Llobet A, Spring DR (2010) Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun 1(80):1–13
Giannetti AM (2011) From experimental design to validated hits a comprehensive walk-through of fragment lead identification using surface plasmon resonance. Methods Enzymol 493:169–218
Gomes NGM, Pereira DM, Valentão P et al (2018) Hybrid MS/NMR methods on the prioritization of natural products: application in drug discovery. J Pharm Biomed Anal 147:234–249
Goodford PJ (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 28:849–857
Gossert AD, Jahnke W (2016) NMR in drug discovery: a practical guide to identification and validation of ligands interacting with biological macromolecules. Prog Nucl Magn Reson Spectrosc 97:82–125
Gribbon P, Sewing A (2005) High-throughput drug discovery: what can we expect from HTS? Drug Discov Today 10:17–22
Hajduk PJ (2006) Fragment-based drug design: how big is too big? J Med Chem 49:6972–6976
Hajduk PJ (2011) Small molecules, great potential. Nature 470:42
Hajduk PJ, Greer J (2007) A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov 6:211–219
Hajduk PJ, Huth JR, Fesik SW (2005) Druggability indices for protein targets derived from NMR-based screening data. J Med Chem 48:2518–2525
Hall RJ, Mortenson PN, Murray CW (2014) Efficient exploration of chemical space by fragment-based screening. Prog Biophys Mol Biol 116:82–91
Hann MM, Leach AR, Harper G (2001) Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci 41:856–864
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14:111–129
Henrich CJ, Beutler JA (2013) Matching the power of high throughput screening to the chemical diversity of natural products. Nat Prod Rep 30:1284–1298
Higueruelo AP, Jubb H, Blundell TL (2013) Protein-protein interactions as druggable targets: recent technological advances. Curr Opin Pharmacol 13:791–796
Hiroaki H (2013) Recent applications of isotopic labeling for protein NMR in drug discovery. Expert Opin Drug Discov 8:523–536
Huang R, Leung IKH (2019) Protein-small molecule interactions by WaterLOGSY. Methods Enzymol 615:477–500
Hubbard RE (1997) Can drugs be designed? Curr Opin Biotechnol 8:696–700
Hubbard RE (2005) 3D structure and the drug-discovery process. Mol Biosyst 1:391–406
Hubbard RE (2008) Fragment approaches in structure-based drug discovery. J Synchrotron Radiat 15:227–230
Hubbard RE (2015) The fragment based lead discovery (chapter 5). In: Davis A, Ward SE (eds) The handbook of medicinal chemistry: principles and practices. Wiley, New York, pp 122–153
Hubbard RE (2016) The role of fragment-based discovery in lead finding. In: Erlanson DA, Jahnke W (eds) Chapter 1 in fragment-based discovery. Wiley, New York, pp 3–36
Hubbard RE, Murray JB (2011) Experiences in fragment-based lead discovery. Methods Enzymol 493:509–531
Huber W, Mueller F (2006) Biomolecular interaction analysis in drug discovery using surface plasmon resonance technology. Curr Pharm Des 12:3999–4021
Jencks WP (1981) On the attribution and additivity of binding energies. Proc Natl Acad Sci USA 78:4046–4050
Johnson CN, Erlanson DA, Jahnke W et al (2018) Fragment-to-lead medicinal chemistry publications in 2016. J Med Chem 61:1774–1784
Keserű GM, Hann MM (2017) What is the future for fragment-based drug discovery? Future Med Chem 9:1457–1460
Keserű GM, Erlanson DA, Ferenczy GG et al (2016) Design principles for fragment libraries: maximizing the value of learnings from pharma fragment-based drug discovery (FBDD) programs for use in academia. J Med Chem 59:8189–8206
Kutchukian PS, Wassermann AM, Lindvall MK et al (2015) Large scale meta-analysis of fragment-base screening campaigns: privileged fragments and complementary technologies. J Biomol Screen 20:588–596
Lamoree B, Hubbard RE (2017) Current perspectives in fragment-based lead discovery (FBLD). Essays Biochem 61:453–464
Lanz J, Riedl R (2015) Merging allosteric and active site binding motifs: de novo generation of target selectivity and potency via natural-product-derived fragments. Chem Med Chem 10:451–454
Leach AR, Hann MM, Burrows JN et al (2006) Fragment screening: an introduction. Mol Biosyst 2:430–446
Lipinski C, Hopkins A (2004) Navigating chemical space for biology and medicine. Nature 432:855–861
Long C, Sauleau P, David B et al (2003) Bioactive flavonoids of Tanacetum parthenium revisited. Phytochemistry 64:567–569
Long C, Beck J, Cantagrel F et al (2012) Proteasome inhibitors from Neoboutonia melleri. J Nat Prod 75:34–47
Lovering F, Bikker J, Humblet C (2009) Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem 32:6752–6756
Maybridge (2019) The Maybridge Ro3 2500 Diversity Fragment Library. https://www.maybridge.com/portal/alias__Rainbow/lang__en/tabID__230/DesktopDefault.aspx. Cited 14 Mar 2019
Mayer M, Meyer B (1999) Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed Engl 38:1784–1788
Montfort RLM, Workman P (2017) Structure-based drug design: aiming for a perfect fit. Essays Biochem 61:431–437
Morgan P, Brown DG, Lennard S et al (2018) Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nat Rev Drug Discov 17:167–181
Morley AD, Pugliese A, Birchall K et al (2013) Fragment-based hit identification: thinking in 3D. Drug Discov Today 18:1221–1227
Mullin R (2004) Drug discovery: as high-throughput screening draws fire, researchers leverage science to put automation into perspective. Chem Eng News 82:23–32
Murray CW, Rees D (2009) The rise of fragment-based drug discovery. Nat Chem 1:187–192
Murray CW, Verdonck ML, Rees DC (2012) Experiences in fragment-based drug discovery. Trends Pharm Sci 33:224–232
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661
Oganesyan I, Lento C, Wilson DJ (2018) Contemporary hydrogen deuterium exchange mass spectrometry. Methods 144:27–42
Oprea TI (2002) Current trends in lead discovery: are we looking for the appropriate properties? J Comput Aided Mol Des 16:325–334
Over B, Wetzel S, Grütter C et al (2013) Natural-product-derived fragments for fragment-based ligand discovery. Nat Chem 5:21–28
Pahl A, Waldmann H, Kumar K (2017) Exploring natural product fragments for drug and probe discovery. Chimia 71:653–660
Pascolutti M, Quinn RJ (2014) Natural products as lead structures: chemical transformations to create lead-like libraries. Drug Discov Today 19:215–221
Pascolutti M, Campitelli M, Nguyen B et al (2015) Capturing nature’s diversity. PLoS ONE 10(4):e0120942
Paterson I, Anderson EA (2005) The renaissance of natural products as drug candidates. Science 310:451–453
Pedro L, Quinn RJ (2016) Native mass spectrometry in fragment-based drug discovery. Molecules 21:984–999
Pouny I, Batut M, Vendier L et al (2014) Cytisine-like alkaloids from Ormosia hosiei Hemsl. & E.H. Wilson. Phytochemistry 107:97–101
Prescher H, Koch G, Schuhmann T, Ertl P (2017) Construction of a 3D-shaped, natural product like fragment library by fragmentation and diversification of natural products. Bioorg Med Chem 25:921–925
Price AJ, Howard S, Cons BD (2017) Fragment-based drug discovery and its application to challenging drug targets. Essays Biochem 61:475–484
Pye CR, Bertin MJ, Lokey RS et al (2017) Retrospective analysis of natural products provides insights for the future discovery trends. Proc Natl Acad Sci USA 114:5601–5606
Rachman MM, Barril X, Hubbard RE (2018) Predicting how drug molecules bind to their protein targets. Curr Opin Pharmacol 42:34–39
Rees DC, Congreve M, Murray CW et al (2004) Fragment-based lead discovery. Nat Rev Drug Discov 3:660–672
Renaud JP, Chung CW, Danielson UH et al (2016) Biophysics in drug discovery: impact, challenges and opportunities. Nat Rev Drug Discov 15:679–698
Rodrigues T, Reker D, Kunze J et al (2015) Revealing the macromolecular targets of fragment-like natural products. Angew Chem Int Ed Engl 127:10662–10666
Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754
Romasanta AKS, van der Sijde P, Hellsten I et al (2018) When fragments link: a bibliometric perspective on the development of fragment-based drug discovery. Drug Discov Today 23:1596–1609
Roughley SD, Hubbard RE (2011) How well can fragments explore accessed chemical space? A case study from heat shock protein 90. J Med Chem 54:3989–4005
Scannel JW, Blanckley A, Boldon H et al (2012) Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov 11:191–200
Schulz MN, Hubbard RE (2009) Recent progress in fragment-based lead discovery. Curr Opin Pharmacol 9:615–621
Schulz MN, Landström J, Bright K et al (2011) Design of a fragment library that maximally represents available chemical space. J Comput Aided Mol Des 25:611–620
Scott DE, Coyne AG, Hudson SA et al (2012) Fragment-based approaches to drug discovery and chemical biology. Biochemistry 51:4990–5003
Sharma S, Karri K, Thapa I et al (2016) Identifying enriched drug fragments as possible candidates for metabolic engineering. BMC Med Genom 9(Suppl 2):167–177
Shoichet BK (2013) Nature’s pieces. Nat Chem 5:9–10
Shuker SB, Hajduk PJ, Meadows RP et al (1996) Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531–1534
Siegel MG, Vieth M (2007) Drugs in other drugs: a new look at drugs as fragments. Drug Discov Today 12:71–79
Speck-Planche A (2018) Recent advances in fragment-based computational drug design: tackling simultaneous targets/biological effects. Future Med Chem 10:2021–2024
Sugiki T, Furuita K, Fujiwara T, Kojima C (2018) Current NMR techniques for structure-based drug discovery. Molecules 23:E148. https://doi.org/10.1007/s10930-018-9797-3
Valenti D, Hristeva S, Tzalis D et al (2019) Clinical candidates modulating protein–protein interactions: the fragment-based experience. Eur J Med Chem 167:76–95
Veber DF, Johnson SR, Cheng HY et al (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623
Velvadapu V, Farmer BT, Reitz AB (2015) Fragment-based drug discovery. In: Wermuth C, Aldous D, Raboisson P, Rognan D (eds) Chapter 7 in the practice of medicinal chemistry, 4th edn. Academic Press, London
Verlinde CL, Rudenko G, Hol WG (1992) In search of new lead compounds for trypanosomiasis drug design: a protein structure-based linked-fragment approach. J Comput Aided Mol Des 6:131–147
Viegas A, Manso J, Nobrega FL et al (2011) Saturation-transfer difference (STD) MMR: a simple and fast method for ligand screening and characterization of protein binding. J Chem Educ 88:990–994
Vivat Hannah V, Atmanene C, Zeyer D, Van Dorsselaer A, Sanglier-Cianférani S (2010) Native MS: an ‘ESI’ way to support structure- and fragment-based drug discovery. Future Med Chem 2:35–50
Vu H, Roullier C, Campitelli M et al (2013) Plasmodium gametocyte inhibition identified from a natural-product-based fragment library. ACS Chem Biol 8:2654–2659
Vu H, Pedro L, Mak T et al (2018) Fragment-based screening of a natural product library against 62 potential malaria drug targets employing native mass spectrometry. ACS Infect Dis 4:431–444
Williams G, Ferenczy GG, Ulander J et al (2017) Binding thermodynamics discriminates fragments from druglike compounds: a thermodynamic description of fragment-based drug discovery. Drug Discov Today 22:681–689
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
David, B., Grondin, A., Schambel, P. et al. Plant natural fragments, an innovative approach for drug discovery. Phytochem Rev 19, 1141–1156 (2020). https://doi.org/10.1007/s11101-019-09612-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11101-019-09612-4