Abstract
Neuropeptides are derived from large and inactive proteins which require endoproteolytic processing for the biosynthesis of the bioactive peptides. The maturation of pro-neuropeptide to neuropeptide is believed to be performed by ortholog pro-protein convertase EGL-3 in Caenorhabditis elegans (C. elegans). Furthermore, ortholog of Cathepsin L, CPL-1 are found in C. elegans and can potentially cleave paired basic amino acids at the N-terminal suggesting the presence of both pathways. The objective of this study was to decipher the role of EGL-3 in the proteolysis of FMRF amide-related peptides (FLPs) or neuropeptide-like proteins (NLPs) using synthetic surrogate peptides based on a universal enzymatic cleavage pattern published by Schechter and Berger and used widely in enzymology. The results show evidence that proteolysis controls FLP-21 and NLP-8 related neuropeptide levels in C. elegans. Surrogate peptides were degraded rapidly when exposed to C. elegans S9 fractions leading to the formation of specific peptide fragments related to EGL-3 and CPL-1 pathway. The results suggest that CPL-1 pathway does not compensate for the loss of the EGL-3 pathway. Proteolysis of pro-neuropeptides associated to FLP-21 and NLP-8 in elg-3 mutants are severely hampered leading to a lack of mature bioactive neuropeptides.

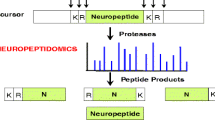
(Reproduced with permission from Hook et al. [6]). b FLP-21 processing by proprotein convertases EGL-3 leading to active FLP-21 neuropeptide. c NLP-8 processing by proprotein convertases EGL-3 and carboxypeptidase E EGL-21 leading to several actives neuropeptides. Identification of surrogate peptides used for further in vitro exploration was based on subsite nomenclature adopted from a scheme developed by Schechter and Berger [26, 27] and used to describe of enzyme specificities






Similar content being viewed by others
References
Hook V, Brennand KJ, Kim Y, Toneff T, Funkelstein L, Lee KC et al (2014) Human iPSC neurons display activity-dependent neurotransmitter secretion: aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep 3(4):531–538
Rouillé Y, Duguay SJ, Lund K, Furuta M, Gong Q, Lipkind G et al (1995) Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front Neuroendocrinol 16(4):322–361
Seidah NG, Day R, Marcinkiewicz M, Chretien M (1998) Precursor convertases: an evolutionary ancient, cell-specific, combinatorial mechanism yielding diverse bioactive peptides and proteins. Ann N Y Acad Sci 839(1):9–24
Alberts B, Johnson A, Lewis J et al (2002) Molecular biology of the cell, 4th edn. Garland Science, New York
Steiner DF, Smeekens SP, Ohagi S, Chan SJ (1992) The new enzymology of precursor processing endoproteases. J Biol Chem 267:23435–23438
Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol 48:393
Hook VY (2006) Protease pathways in peptide neurotransmission and neurodegenerative diseases. Cell Mol Neurobiol 26(4–6):447–467
Seidah N, Prat A (2002) Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem 38:79–94
Scamuffa N, Calvo F, Chrétien M, Seidah NG, Khatib AM (2006) Proprotein convertases: lessons from knockouts. FASEB J 20(12):1954–1963
Hwang SR, O’Neill A, Bark S, Foulon T, Hook V (2007) Secretory vesicle aminopeptidase B related to neuropeptide processing: molecular identification and subcellular localization to enkephalin-and NPY-containing chromaffin granules. J Neurochem 100(5):1340–1350
Zhang X, Pan H, Peng B, Steiner DF, Pintar JE, Fricker LD (2010) Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J Neurochem 112(5):1168–1179
Minokadeh A, Funkelstein L, Toneff T, Hwang SR, Beinfeld M, Reinheckel T, Hook V (2010) Cathepsin L participates in dynorphin production in brain cortex, illustrated by protease gene knockout and expression. Mol Cell Neurosci 43(1):98–107
Saidi M, Kamali S, Ruiz AO, Beaudry F (2015) Tachykinins processing is significantly impaired in PC1 and PC2 mutant mouse spinal cord S9 fractions. Neurochem Res 40(11):2304–2316
Salem JB, Nkambeu B, Beaudry F (2018) Characterization of neuropeptide K processing in rat spinal cord S9 fractions using high-resolution quadrupole–orbitrap mass spectrometry. Biomed Chromatogr 32(6):e4204
Orduna AR, Beaudry, F (2016) Characterization of endoproteolytic processing of dynorphins by proprotein convertases using mouse spinal cord S9 fractions and mass spectrometry. Neuropeptides 57:85–94
Mills H, Wragg R, Haplak V, Castelleto M, Zahratka J, Harris G, Summers P, Korchnak A, Law W, Bamber B, Komuniecki R (2012) Monoamines and neuropeptides interact to inhibit aversive behaviour in Caenorhabditis elegans. EMBO J 31(3):667–678
Li C (2005) The ever-expanding neuropeptide gene families in the nematode Caenorhabditis elegans. Parasitology 131(Suppl):S109–S127
Hu Z, Pym ECG, Babu K, Vashlishan-Murray AB, Kaplan JM (2011) A neuropeptide-mediated stretch response links muscle contraction to changes in neurotransmitter release. Neuron 71:92–102
Glauser DA, Chen WC, Agin R, MacInnis B, Hellman AB, Garrity PA, Man-WahTan, Goodman MB (2011) Heat avoidance is regulated by Transient Receptor Potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genet Soc Am 188:91–103
Komuniecki R, Harris G, Haplak V (2012) Monoamines activate neuropeptide signaling cascades to modulate nociception in C. elegans: a useful model for the modulation of chronic pain? Invertebr Neurosci 12:53–61
Kass J, Jacob TC, Kim P, Kaplan JM (2001) The EGL-3 pro-protein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci 21(23):9265–9672
Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM (2013) Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron 78(5):869–880
Husson SJ, Clynen E, Baggerman G, Janssen T, Schoofs L (2006) Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J Neurochem 98(6):1999–2012
Husson SJ, Janssen T, Baggerman G, Bogert B, Kahn-Kirby AH, Ashrafi K, Schoofs L (2007) Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J Neurochem 102(1):246–260
Wittenburg N, Baumeister R (1999) Thermal avoidance in Caenorhabditis elegans: an approach to the study of nociception. PNAS 96(18):10477–10482
Schechter I, Berger A (1967) On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 27(2):157–162
Schechter I, Berger A (1968) On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem Biophys Res Commun 32(5):898–902
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94
Margie O, Palmer C, Chin-Sang I (2013) C. elegans chemotaxis assay. J Vis Exp 74:1–6
Roepstorff P, Fohlman J (1984) Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom 11(11):601
Bousquet-Moore D, Mains RE, Eipper BA (2010) PAM and copper—a gene/nutrient interaction critical to nervous system function. J Neurosci Res 88(12):2535–2545
Metaxakis A, Petratou D, Tavernarakis N (2018) Multimodal sensory processing in Caenorhabditis elegans. Open Biol. 8(6):180049
Ma L, Zhao Y, Chen Y, Cheng B, Peng A, Huang K (2018) Caenorhabditis elegans as a model system for target identification and drug screening against neurodegenerative diseases. Eur J Pharmacol 819:169–180
Blein-Nicolas M, Zivy M (2016) Thousand and one ways to quantify and compare protein abundances in label-free bottom-up proteomics. Biochim Biophys Acta 1864(8):883–895
Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, Van Sluyter SC, Haynes PA (2011) Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11:535–553
Acknowledgements
This project was funded by the National Sciences and Engineering Research Council of Canada (F. Beaudry discovery Grant No. RGPIN-2015-05071). The mass spectrometry analyses were performed using an infrastructure funded by the Canadian Foundation for Innovation (CFI) and the Fonds de Recherche du Québec (FRQ), Government of Quebec (F. Beaudry CFI John R. Evans Leaders Grant No. 36706). A PhD scholarship was awarded to J. Ben Salam with a grant obtained from Fondation de France (DN Arvanitis Grant No. RAF18002BBA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Salem, J.B., Nkambeu, B., Arvanitis, D.N. et al. Deciphering the Role of EGL-3 for Neuropeptides Processing in Caenorhabditis elegans Using High-Resolution Quadrupole–Orbitrap Mass Spectrometry. Neurochem Res 43, 2121–2131 (2018). https://doi.org/10.1007/s11064-018-2636-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-018-2636-2