Abstract
Purpose
Isocitrate dehydrogenase (IDH) and 1p19q codeletion status are importantin providing prognostic information as well as prediction of treatment response in gliomas. Accurate determination of the IDH mutation status and 1p19q co-deletion prior to surgery may complement invasive tissue sampling and guide treatment decisions.
Methods
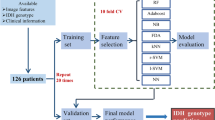
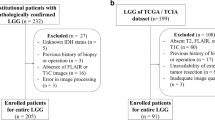
Preoperative MRIs of 538 glioma patients from three institutions were used as a training cohort. Histogram, shape, and texture features were extracted from preoperative MRIs of T1 contrast enhanced and T2-FLAIR sequences. The extracted features were then integrated with age using a random forest algorithm to generate a model predictive of IDH mutation status and 1p19q codeletion. The model was then validated using MRIs from glioma patients in the Cancer Imaging Archive.
Results
Our model predictive of IDH achieved an area under the receiver operating characteristic curve (AUC) of 0.921 in the training cohort and 0.919 in the validation cohort. Age offered the highest predictive value, followed by shape features. Based on the top 15 features, the AUC was 0.917 and 0.916 for the training and validation cohort, respectively. The overall accuracy for 3 group prediction (IDH-wild type, IDH-mutant and 1p19q co-deletion, IDH-mutant and 1p19q non-codeletion) was 78.2% (155 correctly predicted out of 198).
Conclusion
Using machine-learning algorithms, high accuracy was achieved in the prediction of IDH genotype in gliomas and moderate accuracy in a three-group prediction including IDH genotype and 1p19q codeletion.


Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ostrom QT et al (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 16(7):896–913
Bi WL, Beroukhim R (2014) Beating the odds: extreme long-term survival with glioblastoma. Neuro Oncol 16(9):1159–1160
Recht LD, Lew R, Smith TW (1992) Suspected low-grade glioma: is deferring treatment safe? Ann Neurol 31(4):431–436
Parsons DW et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897):1807–1812
Eckel-Passow JE et al (2015) Glioma Groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372(26):2499–2508
Hartmann C et al (2010) Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 120(6):707–718
Houillier C et al (2010) IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75(17):1560–1566
Louis DN et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Weller M et al (2013) Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol 14(9):e370–e379
SongTao Q et al (2012) IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 103(2):269–273
Molenaar RJ et al (2015) Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant inhibitor AGI-5198. Cancer Res 75(22):4790–4802
Mohrenz IV et al (2013) Isocitrate dehydrogenase 1 mutant R132H sensitizes glioma cells to BCNU-induced oxidative stress and cell death. Apoptosis 18(11):1416–1425
Lowery MA et al (2017) Phase I study of AG-120, an IDH1 mutant enzyme inhibitor: results from the cholangiocarcinoma dose escalation and expansion cohorts. J Clin Oncol 35(15_suppl):4015–4015
Mellinghoff IK et al. (2016) ACTR-46. AG120, a first-in-class mutant IDH1 inhibitor in patients with recurrent or progressive IDH1 mutant glioma: results from the phase 1 glioma expansion cohorts. Oxford University Press, Oxford
Erdem-Eraslan L et al (2013) Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol 31(3):328
Cairncross G et al (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31(3):337
Tang L et al (2018) Reduced expression of DNA repair genes and chemosensitivity in 1p19q codeleted lower-grade gliomas. J Neuro-Oncol. https://doi.org/10.1007/s11060-018-2915-4
Brat DJ et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498
Kickingereder P et al (2015) IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 5:16238
Biller A et al (2016) Improved brain tumor classification by sodium MR imaging: prediction of IDH mutation status and tumor progression. AJNR Am J Neuroradiol 37(1):66–73
Pope WB et al (2012) Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol 107(1):197–205
Andronesi OC et al (2012) Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med 4(116):116ra4
Choi C et al (2012) 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 18(4):624–629
Stadlbauer A et al (2017) MR imaging-derived oxygen metabolism and neovascularization characterization for grading and IDH gene mutation detection of gliomas. Radiology 283(3):799–809
Iwadate Y et al (2016) Molecular imaging of 1p/19q deletion in oligodendroglial tumours with 11C-methionine positron emission tomography. J Neurol Neurosurg Psychiatry 87:1016–1021
Fellah S et al (2013) Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? Am J Neuroradiol 34(7):1326–1333
Jansen NL et al (2012) Prediction of oligodendroglial histology and LOH 1p/19q using dynamic [18F] FET-PET imaging in intracranial WHO grade II and III gliomas. Neuro-oncology 14(12):1473–1480
Clark K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057
https://wiki.cancerimagingarchive.net/display/Public/TCGA-LGG#0e9decd5f9664ff990f9a9d7e5c43631
Chang K et al (2016) Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol 18(12):1680–1687
Fedorov A et al (2012) 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30(9):1323–1341
Iglesias JE et al (2011) Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 30(9):1617–1634
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
James G et al. (2013) An introduction to statistical learning, vol 112. Springer, New York
Hanley JA, McNeil BJ (1983) A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 148(3):839–843
Eoli M et al (2006) Reclassification of oligoastrocytomas by loss of heterozygosity studies. Int J Cancer 119(1):84–90
Megyesi JF et al (2004) Imaging correlates of molecular signatures in oligodendrogliomas. Clin Cancer Res 10(13):4303–4306
Jenkinson MD et al (2006) Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain 129(Pt 7):1884–1891
Brown R et al (2008) The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res 14(8):2357–2362
Liu C et al (2012) Towards MIB-1 and p53 detection in glioma magnetic resonance image: a novel computational image analysis method. Phys Med Biol 57(24):8393–8404
Metellus P et al (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 120(6):719–729
Qi S et al (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 7(6):1895–1902
Park Y et al (2018) Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. Am J Neuroradiol 39(1):37–42
Zhang B et al (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19(1):109–117
Yu J et al (2017) Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol 27(8):3509–3522
Zhou H et al (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19(6):862–870
Akkus Z et al (2017) Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging 30(4):469–476
Parmar C et al (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087
Fernández-Delgado M et al (2014) Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res 15(1):3133–3181
Acknowledgements
This work was supported by the Natural Science Foundation of China (81301988 to L.Y.), ShenghuaYuying Project of Central South University to L.Y. and the Natural Science Foundation of Hunan Province for Young Scientists, China (Grant No: 2018JJ3709 to Li Yang). This project was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under Award Number 5T32EB1680 to K. Chang. This project was supported by the RSNA research fellow Grant to H.X.B (RF1802), SIR Foundation resident research grant to H.X.B, and Research Fund for International Young Scientist by the National Natural Science Foundation of China (818580410556 to H.X.B.). This research was carried out in whole or in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health.
Funding
This work was supported by the Natural Science Foundation of China (81301988 to L.Y.), ShenghuaYuying Project of Central South University to L.Y. Project supported by the Natural Science Foundation of Hunan Province for Young Scientists, China (Grant No: 2018JJ3709 to Li Yang). This project was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under Award Number 5T32EB1680 to K. Chang. This project was supported by the RSNA research fellow grant to H.X.B (RF1802), SIR Foundation resident research grant to H.X.B, and Research Fund for International Young Scientist by the National Natural Science Foundation of China (818580410556 to H.X.B.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
For this type of study, formal consent is not required.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, H., Chang, K., Bai, H.X. et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J Neurooncol 142, 299–307 (2019). https://doi.org/10.1007/s11060-019-03096-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-019-03096-0