Abstract
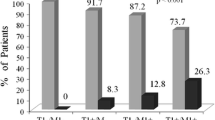
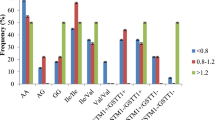
Polymorphism in metabolizing enzymes can influence drug response as well as the risk for adverse drug reactions. Nevertheless, there are still few studies analyzing the consequence of polymorphisms for the Glutathione-S-transferases (GST) gene to drug response in chronic myeloid leukemia (CML). This study reports, the influence of GSTP1*B and GSTT1/GSTM1null polymorphisms in response to imatinib in CML patients in a Brazilian population. One hundred thirty-nine CML patients from the Clinical Hospital of Goiânia, Goiás, Brazil, treated with imatinib were enrolled in this study. Genotyping of GSTT1 and GSTM1 genes deletions were performed by qPCR and of GSTP1 gene was performed by RFLP-PCR. The frequency of GSTP1*1B, GSTT1 and GSTM1null polymorphisms were determined for all patients. The influence of each patient’s genotypes was analyzed with the patient’s response to imatinib treatment. Brazilian CML patients revealed GSTT1 and GSTM1 genes deletions. GSTT1 deletion was found in 19.3% of patients and GSTM1 deletion in 48.7% of patients with CML. GSTT1/GSTM1 deletion was found in 11.7% in Brazilian CML patients. The “G allele” of GSTP1*B, is associated with later cytogenetic response in imatinib therapy. While, the gene presence combined with GG genotype (GSTM1 present/GSTPI-GG) conferred a tend to a later cytogenetic response to patients. GSTP1*B and GSTT1/GSTM1null polymorphisms influence treatment response in CML. Brazilian CML patients presenting GSTP1 AA/AG genotypes alone and in combination with GSTT1 null reach the cytogenetic response faster, while patients presenting GSTP1-GG and GSTMI positive genotypes may take longer to achieve cytogenetic response. As a result, it allows a better prognosis, with the use of an alternative therapy, other than reducing treatment cost.



Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66:7–30. https://doi.org/10.3322/caac.21332
Rowley JD (1973) A new consistent chromossomal abnormality in chronic myelogenous leukaemia indetified by quinacrine fluorescence and giemsa staining. Nature 243:290–293. https://doi.org/10.1038/243290a0
Jabbour EJ, Kantarjian H (2014) CME Information: chronic myeloid leukemia: 2014 update. Am J Hematol 89:547–556. https://doi.org/10.1002/ajh.34
Hochhaus A, PLR (2004) Imatinib therapy in chronic myelogenous leukemia: strategies to avoid and overcome resistance ´. Leukemia 18:1321–1331. https://doi.org/10.1038/sj.leu.2403426
Van Etten RA (2007) Oncogenic signaling: new insights and controversies from chronic myeloid leukemia. J Exp Med 204:461–465. https://doi.org/10.1084/jem.20062335
Valent P (2007) Imatinib-resistant chronic myeloid leukemia ( CML ): current concepts on pathogenesis and new emerging pharmacologic approaches. Biol Targets Ther 1:433–448
Okuda K, Weisberg E, Gilliland DG, Griffin JD (2001) ARG tyrosine kinase activity is inhibited by STI571. Blood J 97:2440–2449
Frazer R, Irvine AE, McMullin MF (2007) Chronic myeloid leukaemia in The 21st century. Ulster Med J 76:8–17
Liu ET, Johnston PG (2013) Personalized medicine: does the molecular suit fit? Oncologist 18:653–654. https://doi.org/10.1634/theoncologist.2013-0191
Hanfstein B, Müller MC, Hehlmann R et al (2012) Early molecular and cytogenetic response is predictive for long-term progression-free and overall survival in chronic myeloid leukemia (CML). Leukemia 26:2096–2102. https://doi.org/10.1038/leu.2012.85
Pray LA (2008) Gleevec: the breakthrough in cancer treatment. Nat Educ 1:37. https://doi.org/10.1038/306239a0
Francis S, Lucas C, Lane S et al (2013) A population study showing that the advent of second generation tyrosine kinase inhibitors has improved progression-free survival in chronic myeloid leukaemia. Leuk Res 37:752–758. https://doi.org/10.1016/j.leukres.2013.04.003
Kassogue Y, Quachouh M, Dehbi H et al (2014) Effect of interaction of glutathione S -transferases ( T1 and M1 ) on the hematologic and cytogenetic responses in chronic myeloid leukemia patients treated with imatinib. Med Oncol 7:31–47. https://doi.org/10.1007/s12032-014-0047-z
Terada T, Noda S, Inui K-I (2015) Management of dose variability and side effects for individualized cancer pharmacotherapy with tyrosine kinase inhibitors. Pharmacol Ther 152:125–134. https://doi.org/10.1016/j.pharmthera.2015.05.009
Vaidya S, Ghosh K, Shanmukhaiah CBRV (2015) Genetic variations of hOCT1 gene and CYP3A4 / A5 genes and their association with imatinib response in Chronic Myeloid Leukemia. Eur J Pharmacol 765:124–130. https://doi.org/10.1016/j.ejphar.2015.08.034
Weich N, Ferri C, Moiraghi B et al (2016) GSTM1 and GSTP1 , but not GSTT1 genetic polymorphisms are associated with chronic myeloid leukemia risk and treatment response. Cancer Epidemiol 44:16–21. https://doi.org/10.1016/j.canep.2016.07.008
Valdes R, Yin D (Tyler) (2016) Fundamentals of pharmacogenetics in personalized, precision medicine. Clin Lab Med 36:447–459. https://doi.org/10.1016/j.cll.2016.05.006
Townsend DM, Tew KD (2003) The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 22:7369–7375. https://doi.org/10.1038/sj.onc.1206940
Mcllwain CC, Townsend DM, Tew KD (2006) Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene 25:1639–1648. https://doi.org/10.1038/sj.onc.1209373
Taspinar M, Aydos S, Comez O et al (2008) CYP1A1 , GST gene polymorphisms and risk of chronic myeloid leukaemia. Swiss Med Wkly 138:12–18 https://doi.org/2008/01/smw-12036
Hegazy LA, Mohamad Nohair SAAAE-A (2010) Glutathione S transferase T1 and M1 gene polymorphisms and risk of myeloid leukemias. Med J Cairo Univ 78:67–75. https://doi.org/10.1007/s13277-014-1810-7
Zhou L, Zhu Y-Y, Zhang X-D et al (2013) Risk effects of GST gene polymorphisms in patients with acute myeloid leukemia: a prospective study. Asian Pacific J Cancer Prev 14:3861–3864. https://doi.org/10.7314/APJCP.2013.14.6.3861
He H, Zhang X, Sun J et al (2014) Glutathione S-transferase gene polymorphisms and susceptibility to chronic myeloid leukemia. Tumor Biol 35:6119–6125. https://doi.org/10.1007/s13277-014-1810-7
Kassogue Y, Dehbi H, Quachouh M et al (2015) Association of glutathione S-transferase (GSTM1 and GSTT1) genes with chronic myeloid leukemia. Springerplus 4:3–7. https://doi.org/10.1186/s40064-015-0966-y
Davies A, Giannoudis A, Zhang JE et al (2014) Dual glutathione-S-transferase-θ1 and -μ1 gene deletions determine imatinib failure in chronic myeloid leukemia. Clin Pharmacol Ther 96:694–703. https://doi.org/10.1038/clpt.2014.176
Al-achkar W, Moassass F, Aroutiounian R (2017) Effect of Glutathione S-transferase mu 1 (GSTM1) gene polymorphism on chronic myeloid leukemia risk and Imatinib treatment response. Meta Gene 1. https://doi.org/10.1016/j.mgene.2017.02.006
Rostami G (2019) Influence of glutathione S - transferases ( GSTM1 , GSTT1 , and GSTP1 ) genetic polymorphisms and smoking on susceptibility risk of chronic myeloid leukemia and treatment response. Mol Genet Genomic Med 7(7):e00717. https://doi.org/10.1002/mgg3.717
Lee N, Min S, Jeong P et al (2020) Association between glutathione - S - transferase gene polymorphisms and responses to tyrosine kinase inhibitor treatment in patients with chronic myeloid leukemia: a meta - analysis. Target Oncol 15(1):47–54. https://doi.org/10.1007/s11523-020-00696-z
Skidmore TE (2019) Fact and myth: discovering a racial problem in Brazil. In: Population, ethnicity, and nation-building
Souza CL, Barbosa CG, de Moura Neto JP et al (2008) Polymorphisms in the glutathione S-transferase theta and mu genes and susceptibility to myeloid leukemia in Brazilian patients. Genet Mol Biol 31:39–41. https://doi.org/10.1590/S1415-47572008000100008
Lordelo GS, Akimoto AK, Alves PCZ (2012) Association between methylene tetrahydrofolate reductase and glutathione S-transferase M1 gene polymorphisms and chronic myeloid leukemia in a Brazilian population. Genet Mol Res 11:1013–1026
Vieira-Mion AL, Pereira NF, Funke VAM, Pasquini R (2017) Molecular response to imatinib mesylate of Brazilian patients with chronic myeloid leukemia. Rev Bras Hematol Hemoter 39:210–215. https://doi.org/10.1016/j.bjhh.2017.04.007
Baccarani M, Deininger MW, Rosti G et al (2013) Review Article European LeukemiaNet recommendations for the management of chronic myeloid leukemia : 2013. Blood J 122:872–885. https://doi.org/10.1182/blood-2013-05-501569
Pinheiro DS, Rocha Filho CR, Mundim CA et al (2013) Evaluation of glutathione S-transferase GSTM1 and GSTT1 deletion polymorphisms on Type-2 diabetes mellitus risk. PLoS One 8:1–5. https://doi.org/10.1371/journal.pone.0076262
Marín F, Garcia N, Munoz X, Capella G, González CA, Agudo A, Sala N (2010) Simultaneous genotyping of GSTT1 and GSTM1 null polymorphisms by melting curve analysis in presence of SYBR green I Fa. J Mol Diagnosis 12:300–304. https://doi.org/10.2353/jmoldx.2010.090076
Harries LW, Stubbins MJ, Forman D et al (1997) Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder , testicular and prostate cancer. Carcinogenesis 18:641–644. https://doi.org/10.1093/carcin/18.4.641
Purcell S, Cherny SS, Sham PC (2003) Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 19(1):149–150. https://doi.org/10.1093/bioinformatics/19.1.149
Gschwind H-P, Pfaar U, Waldmeier F et al (2005) Metabolism and disposition of imatinib mesylate in healthy volunteers. Drug Metab Dispos 33:1503–1512. https://doi.org/10.1124/dmd.105.004283.1998
Cho SG, Lee YH, Park HS et al (2001) Glutathione S-transferase Mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J Biol Chem 276:12749–12755. https://doi.org/10.1074/jbc.M005561200
Gate L, Majumdar RS, Lunk A, Tew KD (2004) Increased myeloproliferation in glutathione S-transferase -deficient mice is associated with a deregulation of JNK and Janus kinase/STAT pathways. J Biol Chem 279:8608–8616. https://doi.org/10.1074/jbc.M308613200
Jabbour E (2016) Chronic myeloid leukemia: first-line drug of choice. Am J Hematol 91:59–66. https://doi.org/10.1002/ajh.24249
Vivas W (2008) Manual PRÁTICO DE HEMATOLOGIA, pp 1–33. http://www.aa.med.br/upload/biblioteca/Manual%20de%20Hematologia.pdf
Sim SC, Kacevska M, Ingelman-Sundberg M (2013) Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J 13:1–11
Magno LAV, Talbot J, Talbot T et al (2009) Glutathione S-transferase variants in a Brazilian population. Pharmacology 83:231–236. https://doi.org/10.1159/000205823
Hayes JD, Strange RC (2000) Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 61:154–166. https://doi.org/10.1159/000028396
Sailaja K, Surekha D, Rao DN et al (2010) Association of the GSTP1 gene (Ile105Val) polymorphism with chronic myeloid leukemia. Asian Pacific J Cancer Prev 11:461–464
Tew KD, Manevich Y, Grek C et al (2011) The role of glutathione S-transferase P in signaling pathways and S-glutathionylation in cancer. Free Radic Biol Med 51:299–313. https://doi.org/10.1016/j.freeradbiomed.2011.04.013
Amaral EFL (2009) Caracterização de Migrantes em Goiás e Distrito Federal: 1980–2000. Teor E Soc 17:160–185
Fryer AA, Zhao L, Alldersea J et al (1993) Use of site-directed mutagenesis of allele-specific PCR primers to identify the GSTM1 A, GSTM1 B, GSTM1 A,B and GSTM1 null polymorphisms at the glutathione S-transferase, GSTM1 locus. Biochem J 295:313–315. https://doi.org/10.1042/bj2950313
Mclellan RA, Oscarson M, Alexandrie AK et al (1997) Characterization of a human glutathione S-Transferase μ cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activity. Mol Pharmacol 52(6):958–965. https://doi.org/10.1124/mol.52.6.958
DeJong JL, Mohandas T, Tu C-PD (1991) The human Hb (MU) class Gluthatione S-transferases are encoded by a dispersed gene family. Biochem Biophys Res Commun 180:15–22
Zhong S, Spurr NK, Hayes JD, Wolf CR (1993) Deduced amino acid sequence, gene structure and chromosomal location of a novel human class Mu glutathione S-transferase, GSTM4. Biochem J 291:41–50
Wu W, Peden D, Diaz-Sanchez D (2012) Role of GSTM1 in resistance to lung inflammation. Free Radic Biol Med 53:721–729
Bennour A, Saad A, Sennana H (2016) Chronic myeloid leukemia: relevance of cytogenetic and molecular assays. Crit Rev Oncol Hematol 97:263–274. https://doi.org/10.1016/j.critrevonc.2015.08.020
Marin D, Milojkovic D, Olavarria E et al (2008) European LeukemiaNet criteria for failure or suboptimal response reliably identify patients with CML in early chronic phase treated with imatinib whose eventual outcome is poor. Blood 112:4437–4444. https://doi.org/10.1182/blood-2008-06-162388
Jabbour E, Kantarjian H, O’Brien S et al (2011) The achievement of an early complete cytogenetic response is a major determinant for outcome in patients with early chronic phase chronic myeloid leukemia treated with tyrosine kinase inhibitors. Blood 118:4541–4546. https://doi.org/10.1182/blood-2011-04-348110
Acknowledgments
The authors would like to express their sincere thanks to the employees of the Clinical Hospital—Federal University of Goiás (UFG) for assisting in the collection of the patient data.
Funding
This study was financially supported by the Foundation for Research Support of the State of Goiás (FAPEG) (Grant No. 08/2009 - PPSUS, to Elisângela de Paula Silveira-Lacerda, Ph.D).
Author information
Authors and Affiliations
Contributions
Conceptualization, Kezia Aguiar Delmond, and Elisângela de Paula Silveira-Lacerda.; Methodology, Kezia Aguiar Delmond, Hugo Delleon, AngelaAdamski da Silva Reis, Daniela de Melo e Silva, Elisângela de Paula Silveira-Lacerda.; Software, Kezia Aguiar Delmondand Hugo Delleon.; Validation, Kezia Aguiar Delmond, Hugo Delleon, Daniela de Melo e Silva.; Formal analysis, Kezia Aguiar Delmond, Hugo Delleon, Rebeca Mota Goveiaand Davi Carvalho Abreu.; Investigation, Kezia Aguiar Delmond, Elisângela de Paula Silveira-Lacerda.; resources, Kezia Aguiar Delmond, Adriana do Prado Barbosa, Renato Sampaio Tavares, Carlos Eduardo Anunciação, Elisângela de Paula Silveira-Lacerda.; Data curation, Hugo Delleon, Rebeca Mota Goveia, Thallita Monteiro Teixeira, and Davi Carvalho Abreu.; Writing - Kezia Aguiar Delmond, Elisângela de Paula Silveira-Lacerda.; Writing—review and editing, Kezia Aguiar Delmond, Hugo Delleon, Daniela de Melo e Silva, Francyelli Mello-Andrade, Elisângela de Paula Silveira-Lacerda; Visualization, Kezia Aguiar Delmond, Francyelli Mello-Andrade and Elisângela de Paula Silveira-Lacerda.; Supervision, Elisângela de Paula Silveira-Lacerda.; Project administration, Elisângela de Paula Silveira-Lacerda.; Funding acquisition, Carlos Eduardo Anunciação and Elisângela de Paula Silveira-Lacerda.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest or no financial conflict of interest to disclose.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Ethical Committee of Clinical Hospital of Federal University of Goiás (protocol: 211/2009).
Informed consent
Patients signed their written informed consent, which was approved by the Ethical Committee of Clinical Hospital of Federal University of Goiás (protocol: 211/2009). The informed consent was obtained from the parent(s)/legally autorized representative (LAR)/guardians of the minors involved in the study. Patients or your parent(s)/LAR/guardians signed informed consent regarding publishing their data.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Delmond, K.A., Delleon, H., Goveia, R.M. et al. Influence of genetic polymorphisms in glutathione-S-transferases gene in response to imatinib among Brazilian patients with chronic myeloid leukemia. Mol Biol Rep 48, 2035–2046 (2021). https://doi.org/10.1007/s11033-020-06093-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-06093-z