Abstract
Ventilator-associated pneumonia (VAP) are responsible for an increase in morbidity, mortality, and prolonged hospital stay. A multiplex PCR kit such as the FilmArray® BCID panel could allow early adaptation of antimicrobial therapy, which is crucial for clinical outcomes. The purpose of this study was to test the performances of FilmArray® BCID panel for the detection of bacteria producing VAP. We tested the FilmArray® BCID panel on 50 bronchoalveolar lavages (BALs), from patients hospitalized in two intensive care units at the Angers university hospital, compared to the conventional culture-based method. The sensitivity and the specificity of the FilmArray® BCID panel were 67.2% and 98.9% respectively. They were 88.6% and 98.3% respectively when considering BALs with a positive culture > 104 CFU/mL, and 94.7% and 99.6% respectively if considering BALs with a positive direct examination. This study underlines the good performance of the FilmArray® BCID panel for BAL fluid analysis. In case of positive direct examination, this test allows reliable results that can be obtained at an early stage, facilitating the early adaptation of antimicrobial therapy.
Similar content being viewed by others
Introduction
Ventilator associated pneumonia (VAP) are the main cause of nosocomial infections in intensive care units (ICU) in Europe [1,2,3]. They are responsible for an increase in morbidity with longer hospital stays and have also been associated with an additional financial charge evaluated at almost $40,000 per hospitalization in a group of 88,689 patients in the United States [4]. In addition, this disease is associated with a higher mortality, the mortality attributable to VAP varying from 13 to 30% according to studies [1, 2, 5, 6]. Monitoring, diagnosis, prevention, and treatment of VAP are therefore major challenges for treating patients in ICUs.
However, the diagnosis of VAP is challenging because the clinical signs (fever, change in tracheal secretions, hyperleukocytosis…) are not specific. This means that bacteriological documentation remains essential in order to support the diagnosis, but also to optimize the antimicrobial treatments. Indeed, bacteriological analysis is the cornerstone for the implementation of an appropriate antimicrobial therapy. Several studies have shown that the early introduction of an adapted antibiotic regimen is a key determinant of the outcome of VAP [5, 6]. In addition, it has been shown that rapid bacteriological results allow the de-escalation of broad spectrum empiric therapies [7] and also reduces the use of antibiotics on a large scale [4]. The development of tools providing rapid identifications of the pathogens involved in this disease, in comparison with slower conventional bacteriological culture techniques (48–72 h), may represent a new option to adapt a probabilistic antibiotic treatment as soon as possible. Over the last years, the multiplex PCR techniques based on a syndrome-specific approach has revolutionized the diagnosis in microbiology. These commercial kits allows to simultaneously identify several infectious agents (bacteria, yeast, viruses), but also certain resistance genes [8], therefore allowing early clinical management decisions.
The impact of the multiplex PCR kits used on vials of positive blood cultures has been assessed. Positive impacts of these PCR techniques were proven, as early antimicrobial therapy de-escalation in sepsis [9, 10]. Good performances were also observed for these molecular techniques applied on respiratory samples, such as the pneumonia panel in the Curetis® Unyvero System (Curetis, Holzgerlingen, Germany), for patients who were suspected of having developed a VAP [11].
FilmArray® (bioMerieux, Marcy L’Etoile, France) is an automated multiplex PCR system, in which the blood culture identification (BCID) panel can identify 27 targets (19 bacteria, 5 yeasts and 3 resistance genes). The composition of the FilmArray® BCID panel could be potentially usable in diagnosing VAP in about an hour, to allow an early adaptation of antibiotic treatment. The main bacteria reported as being responsible for VAP (Enterobacteriaceae, Staphylococcus aureus, Pseudomonas aeruginosa, Acinetobacter baumanii, Streptococcus pneumoniae and Haemophilus influenzae) are included in the BCID panel.
The aim of this study is to evaluate the performances of the FilmArray® BCID panel for the diagnosis of bacteria associated with VAP on bronchoalveolar lavage (BAL) liquids sampled from patients hospitalized in intensive care units of a university hospital.
Materials and methods
Type of study
This prospective, monocentric study has been approved by the ethics committee of the Angers University Hospital (CHU Angers), France (Approval No. 2017-61). An informative letter about the study was given to the patient or a family member if the patient was unable to read it.
Organization of the study
The study was conducted from December 2017 to April 2018 in the bacteriology laboratory at Angers University Hospital. Fifty BALs intended for diagnosis were included. These ones were collected beforehand in a medical intensive care unit (24 beds) and in a surgical intensive care unit (12 beds). Specimens meeting the following inclusion criteria were included: BALs recovered from patients intubated and ventilated for at least 48 h, and with a rate of recovery of BAL (volume of saline solution recovered/volume of saline instilled) higher than 10%. Patients with a tracheotomy and who are intubated in the long term were excluded from the study. During the study, clinical and biological data, such as body temperature, number of leukocytes in blood counts, the quantity and appearance of tracheal secretions, the arterial partial pressure of O2 (PaO2), and the fraction of inspired oxygen (FiO2), were collected. Thoracic imagery reports were also taken into account. These data were used to establish the mCPIS score (modified Clinical Pulmonary Infection Score) defined by Luna et al. [12]. The mCPIS is a score evaluating the temperature, the blood leucocyte count, tracheal secretions, the oxygenation and the chest radiograph, with points attributed according to the results of each criteria. A mCPIS > 6 has been commonly used in the literature as a predictor of VAP [13]. Thus, a mCPIS score ≥ 6 measured on the day of the BAL sample were considered as being affected by VAP. Lastly, the other data, such as the presence or absence of antibiotic treatment in the days preceding the BAL sample, previous infection during the stay in the ICU, and the period between intubation and the BAL sample, were also collected.
FilmArray® tests
A FilmArray® BCID test (bioMérieux, Marcy l’Etoile, France) was carried out as recommended by the supplier instructions for blood cultures, using 200 µL of BAL fluid without centrifugation or treatment.
Conventional microbiological techniques
The conventional techniques include BAL culture, as well as a direct examination including a Gram stain and a rapid May-Grünwald-Giemsa stain for bacterial and cytologic analysis (RAL 555, RAL-Diagnostics, Montillac, France). Conventional cultures were carried out by inoculating a blood agar with added nalidixic acid and colistin (CNA, Oxoid, Dardilly, France) incubated in anaerobiosis, a chocolate agar (CHOCV, Oxoid) incubated in an atmosphere enriched with 5% CO2, and a chromogenic agar (UTI, Oxoid) incubated in aerobiosis. Several dilutions of the sample, at 10−1, 10−2 and 10−4, were performed for bacterial quantification (CFU/mL). The cultures were interpreted following the recommendations of the European Society for Clinical Microbiology and Infectious Diseases (https://www.escmid.org/) [14]. The bacteria were identified by matrix-assisted laser ionization time-of-flight mass spectrometry (Vitek MS®, bioMérieux, Marcy l’Etoile, France). The antibiotic susceptibility tests were carried out by microdilutions in a liquid environment (Vitek® 2) or by diffusion in an agar environment according to the isolate identified. The results of the antibiotic susceptibility tests were interpreted in accordance with the recommendations of the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/).
Analysis of the results
In this study, we compared exclusively the results concerning bacterial identifications. The results showing a polymorphic flora, potentially a reflection of colonization, were interpreted as negative. The results were compared at the species level; except for the results delivered at the genus level by the FilmArray® BCID test (Enterococcus sp. and Proteus sp.). Therefore, we limited our comparative analysis to 16 targets (analytes) per sample. A result was considered as true positive (TP) or as true negative (TN) when it totally agreed with the culture. A positive result obtained from the culture and not by the FilmArray® was considered a false negative (FN). In contrast, a result obtained from the FilmArray and not by the culture was considered a false positive (FP). All the discordant results were confirmed by a new test carried out using the FilmArray® BCID. It should be noted that if resistance genes were identified by the FilmArray®, a control was performed on the sample by using the GenXpert® system (Cepheid, Sunnyvale, USA).
The concordance between the two methods was investigated for all BAL fluids. Since the usual positive threshold for the interpretation of BAL fluid cultures is 104 CFU/mL [1], we targeted the concordance analysis on BALs for which at least one potentially pathogenic bacterium has been isolated in culture at a concentration greater than or equal to this threshold. Finally, we examined the agreement by considering BAL fluids with a positive direct examination. When a polymorphic flora without predominance of one or two bacterial forms was observed, direct examination was considered negative because non-contributory for the diagnosis.
Statistics
The data were collected by using EPI-INFO 7.2 software (Center for Disease Control, Atlanta, USA). The statistical analyses (Cohen’s kappa test) were carried out on the Vassar Stats website (http://vassarstats.net). A kappa coefficient between 0.81 and 1 indicated a very strong agreement between the results of the culture and FilmArray®. A kappa coefficient between 0.61 and 0.8 indicated a strong agreement between the two results. Conversely, a kappa coefficient of < 0.61 meant that the two results were not sufficiently in agreement.
The sensitivity (Se) was calculated as 100 × [TP/(TP + FN)] and the specificity (Sp) was calculated as 100 × [TN/(TN + FP)]. The positive predictive value (PPV) was calculated as 100 × [TP/(TP + FP)]. The negative predictive value (NPV) was calculated as 100 × [TN/(TN + FN)]. The 95% confidence intervals were calculated according to the Wilson score method.
Results
Study population
Forty-five patients were included in the study, corresponding to 50 BALs. Two BALs were performed for three patients and three BALs for one patient. Twenty-five patients were hospitalized in the intensive care unit and 20 in the surgical intensive care unit. The average age of the patients was 63 ± 17 years old with a sex ratio (M/F) of 2.46. The most frequent reasons for admission to the intensive care unit were acute respiratory distress syndrome (ARDS) (31%), sepsis (11%), a neurological injury (11%), and multiple trauma (9%). Sixteen (36%) patients had an extrapulmonary infection before their BAL sample. Twenty (45%) patients included in the study had received antibiotic treatment in the 3 days before the BAL sample. The average mCPIS score was 6.4 ± 1.7 (extremes, 3–9). The clinical data is presented in Table 1.
Results obtained from culture and FilmArray® BCID
Culture
Of the 50 BALs analyzed for the study, 30 (60%) showed a positive direct examination as well as an average percentage of infected cells of 6.5% (0–75%), of which 16 BALs (32%) had more than 5% infected cells. Thirty-five BALs (70%) showed a positive culture including at least one pathogenic bacterium with a count in a range from 102 to more than 106 CFU/mL. Fifty-five bacteria were identified by using conventional culture techniques, including 33% of Gram-positive bacteria with a predominance of Staphylococcus aureus (72%) and 67% of Gram-negative bacteria with a predominance of Enterobacteriaceae (51%) and Pseudomonas aeruginosa (30%). Only one isolate within the 13 Staphylococcus aureus isolated by culture was resistant to methicillin. A strain of Klebsiella pneumoniae produced an extended-spectrum beta-lactamase (ESBL) and a strain of Pseudomonas aeruginosa was resistant to ceftazidime and susceptible to imipenem.
FilmArray®
The FilmArray® BCID panel test was positive for 34 (68%) of the 50 BALs analyzed, allowing the identification of 44 bacteria: 39% of Gram-positive bacteria with a predominance of Staphylococcus aureus (59%) and 61% of Gram-negative bacteria with Pseudomonas aeruginosa (37%), Haemophilus influenzae (37%) and Enterobacteriaceae (26%). Three mecA genes were detected by the FilmArray®. However, only one was confirmed by the GenXpert® system.
Comparison
Overall, 800 analytes were tested on the 50 BALs (16 analytes per sample). Overall agreement between the FilmArray® and conventional culture results was 96.6% (773/800). For the FilmArray®, we observed an overall sensitivity of 67.2% (CI 95%: 53.5–78.6%), as well as a specificity of 98.9% (CI 95%: 97.8–99.5%). Thirty-nine tests were positive with the FilmArray® and by culture. The overall PPV was 82.9% (CI 95%: 68.7–91.9%).
We observed 734 negative targets with the FilmArray®, corresponding to an absence of growth by culture of bacteria corresponding to these targets. The overall NPV was 97.5% (CI 95%: 96.0–98.4%). The level of agreement between the results of the culture and FilmArray® was considered to be good with a kappa coefficient of 0.73 (CI 95%: 0.63–0.82). The results of the overall comparison are presented in Table 2.
By limiting the comparison of the two methods to BAL fluids with a culture greater than 104 CFU/mL, the overall agreement was 97.9% (783/800) with a sensitivity of 88.6% (CI 95%: 72.3–96.3%), a specificity of 98.3% (CI 95%: 97.0–99.1%), a PPV of 70.5% (CI 95%: 54.6–82.8%), and a NPV of 99.5% (CI 95%: 98.5%–99.8%). A kappa coefficient of 0.77 (CI 95%: 0.67–0.88) was obtained for the agreement between the conventional cultures (bacteria > 104 UFC/mL) and the results of the FilmArray® BCID panel. The sensitivity of identifications obtained for each bacterium was 100% by excluding the bacteria isolated in the culture but not included in the panel (three Citrobacter sp. and one Acinetobacter radioresistens). The specificity of different bacterial identifications varies between 87% (Haemophilus influenzae) and 100% (Klebsiella pneumoniae).
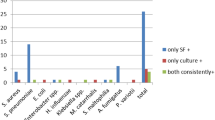
The results of the comparison, taking into consideration the threshold of ≥ 104 CFU/mL, are presented in Table 3. Among the 17 discrepant results out of the 800 targets tested, 13 corresponded to a positive FilmArray® test and a negative culture test (considered FP). The other four contradictions corresponded to a negative FilmArray® result and a positive culture (considered FN) with bacteria not included in the BCID panel. Of the 13 False Positive results, 5 (3 Staphylococcus aureus, 1 Proteus sp. and 1 Enterococcus sp.) corresponded to bacteria isolated in culture at a concentration of less than 104 CFU/mL and 3 (2 Streptococcus pneumoniae and 1 Haemophilus influenzae) of the 13 false positive targets were isolated from the samples taken from the patients treated with antibiotics active on those bacteria. The Fig. 1 presents an overview of discrepancies between the FilmArray® and culture tests, limited to BAL fluids for which at least one bacterium has been identified at a concentration greater than 104 CFU/mL.
Investigation of discrepancies between FilmArray BCID® and culture (> 104 CFU/mL). Concordant results are indicated by the blue bars, discordant results with positive FilmArray BCID results and negative culture results by the orange bars, and discordant results with negative FilmArray BCID results and positive culture results by the grey bars. (Color figure online)
By considering only BALs with positive direct examination, good results were obtained with a sensitivity of 94.7% (CI 95%: 71.9–99.7%), a specificity of 97.9% (CI 95%: 94.9%–99.2%), a PPV value of 78.3% (CI 95%: 55.8–91.7%) and a NPV of 99.6% (CI 95%: 97.2–99.9%) compared to conventional cultures.
Discussion
In the context of the development of new molecular diagnostic tools using a syndrome-specific approach, our study aimed to evaluate the performance of the FilmArray® BCID panel with BAL samples in order to contribute to an early diagnosis of VAP. This study shows the good performance of this test in comparison with conventional microbiological cultures. Indeed, our results show an overall sensitivity of 67.2% and a specificity of 98.9%. In another study, Pulido et al. [15] obtained similar results with a sensitivity of 62.9% and a specificity of 100% when using the FilmArray® BCID panel on BAL samples. Micó et al. [16] also obtained the same results for their 15 BALs studied with the FilmArray® BCID, with a sensitivity of 70% and a specificity of 80%.
In comparison with these two studies, the originality of our work lies in considering two contributory variables for the diagnosis of pneumonia with the BAL: the threshold of bacterial count ≥ 104 CFU/mL and the positive direct examination. To compare the multiplex PCR techniques to culture-based methods for the diagnosis of VAP, the use of a threshold seems relevant. Indeed, the bacterial quantification with respect to this threshold can contribute to differentiate bacterial colonization and infection. By applying the usual recognized threshold for the interpretation of BAL cultures (104 CFU/mL), very good performance was recorded with a sensitivity of 88.6%, a specificity of 98.3%, and a negative predictive value of 99.5%. These molecular techniques are expensive. Furthermore, they do not avoid the culture of BALs, as antibiotic susceptibility testing is essential for the management of the antimicrobial therapy. Thus, the restriction of the use of these techniques to specific clinical situations and their inclusion in a well-defined diagnostic algorithm appear fundamental. In this context, considering the direct examination in such an algorithm seems relevant because the result is obtained very quickly and can contribute to the decision of whether or not to perform a FilmArray® BCID test. It was by considering only BALs with positive direct examination that the best performance was obtained compared to conventional cultures. Therefore, our results would encourage the use of positive direct examination to discriminate the BALs eligible for a rapid diagnostic technique in order to limit the costs and optimize the usefulness of the FilmArray®.
The use of FilmArray® BCID panel for BALs analysis tested in this study presents several limitations. The first limitation is related to the composition of panel, which does not include some Enterobacteriaceae and other bacteria potentially responsible for VAP. This limit is at the origin of the 4 FN identified by considering the threshold of 104 CFU/mL of the culture (1 Acinetobacter radioresistens and 3 Citrobacter sp.). However, the Enterobacteriaceae test of the FilmArray® system was not considered in our analysis despite for the 3 Citrobacter sp. isolated at more than 104 CFU/mL in culture, the Enterobacteriaceae test was positive with the FilmArray®. This result leads us to put these three discrepancies into perspective. Regarding the 13 FP results, 3 positive tests (2 Streptococcus pneumoniae and 1 Haemophilus influenzae) have been recorded from samples of patients treated with antibiotics active on those bacteria. These treatments could explain the absence of culture. Given that the FilmArray® BCID was falsely positive for 5 Haemophilus influenzae tests, it seems not possible to consider tests positive for H. influenzae with this system. These 5 false positive results could be related to a contamination by the operator at the time of the FilmArray® assay. However, the laboratory operates 24/7 with trained laboratory workers [7] and the analytical phase of the FilmArray® system is conducted in a biological safety area with a laminar flow hood. Another possibility is that these false positive results could be linked to low quantity of H. influenzae, that could be not detected by culture methods. This should be investigated by further studies. Another technical limitation is the low number of targets for resistance genes, with in particular the lack of some targets of major interest if we refer to the European epidemiological data (https://ecdc.europa.eu/en/home) (e.g. CTX-M and OXA-48). In addition, we have demonstrated that the detection of the mecA gene by the FilmArray® BCID system did not allow to confirm the presence of MRSA. This may be explained by the presence of methicillin-resistant coagulase-negative staphylococci in the sample.
Overall, our results tend to demonstrate the good performance of multiplex PCR techniques in the rapid diagnosis of VAP. Although it is well established that the introduction of early probabilistic antibiotic therapy reduces the mortality attributable to VAP as compared to a delayed but adapted antimicrobial therapy [5, 17], the prolonged use of broad-spectrum antibiotics promotes the emergence of multi-resistant bacteria [18] The studies conducted by Pailhoriès et al. [7] and Kuti et al. [5] demonstrated that rapid bacteriological documentation during VAP allows the implementation of appropriate antibiotic therapy, often in the form of de-escalation. In addition, the correction of an initially inactive antibiotic therapy is also possible. A bacteriological result by traditional culture method is given at least after a 24 h incubation delay for identification of bacteria in BALs. With the FilmArray® test, the result is available after less than 1 h and a half. For example, a positive Gram-negative bacilli examination will trigger a FilmArray® test. Depending on the outcome, the clinician may adopt either an antibiotic therapy against Pseudomonas aeruginosa or an antibiotic therapy targeted against Enterobacteriaceae. Thus, the appropriate therapy can be initiated within a 2 h delay with this method, in comparison with a 24 h delay with traditional laboratory techniques.
BioMérieux has recently developed a new FilmArray® panel for lower respiratory tract infections. This new test should allow semi quantification and include targets of interest for the search for enzymes responsible for bacterial resistance such as ESBL with the CTX-M gene, and the genes coding for the carbapenemases of the OXA48, KPC, NDM, VIM and IMP types. The question is whether it will really add value to the performance of the BCID FilmArray® test applied to BALs, and at what price. Indeed, the price for the FilmArray® pneumonia test is 210 €, whereas it is 140 € for FilmArray®BCID kit. This last test as it has been shown here can be used for several sample types. Cost-benefit studies will need to be conducted to evaluate it.
References
American Thoracic Society A (2005) Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 171:388–416. https://doi.org/10.1164/rccm.200405-644ST
Melsen WG, Rovers MM, Groenwold RH et al (2013) Attributable mortality of ventilator-associated pneumonia: a meta-analysis of individual patient data from randomised prevention studies. Lancet Infect Dis 13:665–671. https://doi.org/10.1016/S1473-3099(13)70081-1
Hiesmayr M Incidence and attributable mortality of healthcare-associated infections in intensive care units in Europe, 2008–2012. ECDC, Stockholm
Kollef MH, Hamilton CW, Ernst FR (2012) Economic impact of ventilator-associated pneumonia in a large matched cohort. Infect Control Hosp Epidemiol 33:250–256. https://doi.org/10.1086/664049
Kuti EL, Patel AA, Coleman CI Impact of inappropriate antibiotic therapy on mortality in patients with ventilator-associated pneumonia and blood stream infection: a meta-analysis. J Crit Care 23:91–100. https://doi.org/10.1016/j.jcrc.2007.08.007
Iregui M, Ward S, Sherman G et al (2002) Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest 122:262–268
Pailhoriès H, Lemarié C, Kouatchet A et al (2014) The impact of performing bacterial identification and antimicrobial susceptibility testing on bronchoalveolar fluid cultures 24 h a day in a microbiology laboratory. Diagn Microbiol Infect Dis 80:216–221. https://doi.org/10.1016/j.diagmicrobio.2014.07.009
Torres A, Lee N, Cilloniz C et al (2016) Laboratory diagnosis of pneumonia in the molecular age. Eur Respir J 48:1764–1778. https://doi.org/10.1183/13993003.01144-2016
Pardo J, Klinker KP, Borgert SJ et al (2016) Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis 84:159–164. https://doi.org/10.1016/j.diagmicrobio.2015.10.023
Banerjee R, Teng CB, Cunningham SA et al (2015) Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. https://doi.org/10.1093/cid/civ447
Jamal W, Al Roomi E, AbdulAziz LR, Rotimi VO (2014) Evaluation of Curetis Unyvero, a Multiplex PCR-based testing system, for rapid detection of bacteria and antibiotic resistance and impact of the assay on management of severe nosocomial pneumonia. J Clin Microbiol 52:2487–2492. https://doi.org/10.1128/JCM.00325-14
Luna CM, Blanzaco D, Niederman MS et al (2003) Resolution of ventilator-associated pneumonia: prospective evaluation of the clinical pulmonary infection score as an early clinical predictor of outcome. Crit Care Med 31:676–682. https://doi.org/10.1097/01.CCM.0000055380.86458.1E
Zilberberg M, Shorr A (2010) Ventilator-associated pneumonia: the clinical pulmonary infection score as a surrogate for diagnostics and outcome. Clin Infect Dis 51(S1):S131–S135
Cornaglia G, Courcol R, Herrmann J (2012) European manual of clinical microbiology, 1st edn. European Society for Clinical Microbiology and Infectious Diseases, Basel
Pulido MR, González-Galán V, Fernández Cuenca F et al (2018) Application of the BioFire FilmArray blood culture identification panel for rapid identification of the causative agents of ventilator associated pneumonia. Clin Microbiol Infect. https://doi.org/10.1016/j.cmi.2018.06.001
Micó M, Navarro F, de Miniac D et al (2015) Efficacy of the FilmArray blood culture identification panel for direct molecular diagnosis of infectious diseases from samples other than blood. J Med Microbiol 64:1481–1488. https://doi.org/10.1099/jmm.0.000180
Safdar N, Dezfulian C, Collard HR, Saint S (2005) Clinical and economic consequences of ventilator-associated pneumonia: a systematic review. Crit Care Med 33:2184–2193
Kaki R, Elligsen M, Walker S et al (2011) Impact of antimicrobial stewardship in critical care: a systematic review. J Antimicrob Chemother 66:1223–1230. https://doi.org/10.1093/jac/dkr137
Funding
bioMérieux partially provided reagents for the FilmArray assay.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Ethical approval obtained by the local committee of the University Hospital of Angers (2017-61).
Informed consent
An informed consent was obtained from the patient or a family member if the patient was unable to read the informative letter.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sansot, M., Fradin, E., Chenouard, R. et al. Performance of the extended use of the FilmArray® BCID panel kit for bronchoalveolar lavage analysis. Mol Biol Rep 46, 2685–2692 (2019). https://doi.org/10.1007/s11033-019-04710-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04710-0