Abstract
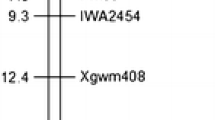
Wheat curl mite (WCM, Aceria tosichella Keifer) and WCM-transmitted wheat streak mosaic virus (WSMV, genus Tritimovirus) are devastating production constraints for wheat in the US Great Plains. Breeding wheat cultivars with genetic resistance to WCM and WSMV is a viable and economically feasible way to reduce yield loss. The objectives of this study were to (a) identify tightly linked markers for WCM resistance in the wheat cultivar TAM 112 (PI 643143) using linkage and association analysis with the 90K Infinium iSelect SNP array and genotyping-by-sequencing, respectively and (b) develop and test kompetitive allele specific PCR (KASP) single-nucleotide polymorphisms (SNPs) for marker-assisted selection (MAS) of WCM resistance. We tested 124 F5:7 recombinant inbred lines (RILs) derived from the cross of TAM 112 and the WCM-susceptible cultivar TAM 111 (PI 631352). All lines were infested with a Texas WCM collection 2 (TWCMC2) that is virulent to resistance found on the wheat-rye 1AL.1RS translocation at the two-leaf stage and were rated for symptoms on the first and second week after infestation. Linkage maps were constructed with 4890 markers, including SNPs, simple sequence repeats (SSRs), and sequence-tagged site (STS) markers covering 21 chromosomes. A WCM resistance gene present in TAM 112 (CmcTAM112) was mapped onto chromosome arm 6DS. A genome-wide association study of wheat streak mosaic (WSM) symptoms from a separate experiment in Colorado showed significant marker-trait associations at the target regions on 6DS where CmcTAM112 was located, which demonstrated the effectiveness of this gene to reduce symptom severity. Four SNPs flanking CmcTAM112 were mapped within 3.6 cM in the biparental mapping population. We developed two KASP markers that are within 1.3 cM distal to CmcTAM112 and tested in diverse germplasm. These two markers can be used in MAS for improving WCM resistance in some wheat genetic backgrounds.


Similar content being viewed by others
References
Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant-Microbe Interact 21:507–517
Andrews JE, Slykhuis JT (1956) Reaction of winter wheat varieties and Triticum × Agropyron hybrids when inoculated with wheat streak mosaic virus by the mite vector Aceria tulipae Keifer. Plant Dis Rep 40:513–516
Appel JA, DeWolf E, Bockus W, Todd T (2012) Preliminary 2011 Kansas wheat disease loss estimates. Kansas Dep. Of Agric., Topeka, KS. http://agriculture.ks.gov/docs/default-source/pp-disease-reports-2012/2012-ks-wheat-disease-loss-estimates.pdf. Accessed 24 Jan 2018
Assanga S, Zhang G, Tan C-T, Rudd JC, Ibrahim A, Xue Q, Chao S, Fuentealba MP, Liu S (2017) Saturated genetic mapping of wheat streak mosaic virus resistance gene Wsm2 in wheat. Crop Sci 57:332–339
Basnet BR, Ibrahim AM, Chen X, Singh RP, Mason ER, Bowden RL, Liu SY, Hays DB, Devkota RN, Subramanian NK, Rudd J (2014) Molecular mapping of stripe rust resistance in hard red winter wheat TAM 111 adapted to the US High Plains. Crop Sci 54:1361–1373
Butler D (2009) asreml: asreml() fits the linear mixed model. R package version 3.0. https://www.vsni.co.uk/downloads/asreml-r/. Accessed 5 Jan 2018
Cakir M, Gupta S, Platz GJ, Ablett GA, Loughman R, Emebiri LC, Poulsen D, Li CD, Lance RCM, Galwey NW, Jones MGK, Appels R (2003) Mapping and validation of gens for resistance to Pyrenophora teres f. teres in barley (Hordeum vulgare L.). Aust J Agric Res 54:1369–1377
Carrera SG, Davis H, Aguirre-Rojas L, Murugan M, Smith CM (2012) Multiple categories of resistance to wheat curl mite (Acari: Eriophidae) expressed in accessions of Aegilops tauschii. J Econ Entomol 105(6):2180–2186
Chapman JA, Mascher M, Buluç A, Barry K, Georganas E, Session A, Strnadova V, Jenkins J, Sehgal S, Oliker L, Schmutz J, Yelick KA, Scholz U, Waugh R, Poland JA, Muehlbauer GJ, Stein N, Rokhsar DS (2015) A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol 16:26
Christian ML, Willis WG (1993) Survival of wheat streak mosaic virus in grass hosts in Kansas from wheat harvest to fall wheat emergence. Plant Dis 77:239–242
Conner RL, Thomas JB, Whelan EDP (1991) Comparison of WCM resistance for control of wheat streak mosaic. Crop Sci 31:315–318
Cox TS, Bockus WW, Gill BS, Sears RG, Harvey TL, Leath S, Brown-Guedira GL (1999) Registration of KS96WGRC40 hard red winter wheat germplasm resistant to wheat curl mite, stagonospora leaf blotch, and septoria leaf blotch. Crop Sci 39:597
Dhakal S, Tan C-T, Paezold L, Fuentealba MP, Rudd JC, Blaser BC, Xue Q, Rush CM, Devkota RN, Liu S (2017) Wheat curl mite resistance in hard red winter wheat in the US Great Plains. Crop Sci 57:53–61
Elshire RJ, Glaubitz JC, Sun Q, Poland J, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:1–10
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. The Plant Genome 4:250–255
Gill BS, Wilson DL, Raupp WJ, Hatchett JH, Cox TS, Amri A, Sears RG (1991a) Registration of KS89WGRC6 Hessian fly-resistant hard red winter wheat germplasm. Crop Sci 31:245
Gill BS, Wilson DL, Raupp WJ, Hatchett JH, Harvey TL, Cox TS, Sears RG (1991b) Registration of KS89WGRC4 hard red winter wheat germplasm with resistance to Hessian fly, greenbug, and soil-borne mosaic virus. Crop Sci 31:246
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler EB (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9(2):e90346. https://doi.org/10.1371/journal.pone.0090346
Goff KE, Ramonell KM (2007) The role and regulation of receptor-like kinases in plant defense. Gene Regul and Syst Bio 1:167–175
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J et al (2012) Phytozyme: a comparative platform for green plant genomics. Nucleic Acids Res 2012:D1178–D1186
Haley SD, Johnson JJ, Peairs FB, Stromberger JA, Heaton EE, Seifert SA, Kottke RA, Rudolph JB, Bai G, Bowden RL, Chen M-S, Chen X, Jin Y, Kolmer JA, Chen R, Seabourn BW (2011) Registration of ‘Snowmass’ wheat. J. Plant Reg. 5:1–4
Haley SD, Johnson JJ, Peairs FB, Stromberger JA, Hudson EE, Seifert SA, Kottke RA, Valdez VA, Rudolph JB, Bai G, Chen X, Bowden RL, Jin Y, Kolmer JA, Chen M-S, Seabourn BW (2012) Registration of ‘Byrd’ wheat. J Plant Reg 6:302–305
Haley SD, Johnson JJ, Peairs FB, Stromberger JA, Hudson EE, Seifert SA, Anderson VA, Rudolph JB, Bai G, Chen X, Bowden RL, Jin Y, Kolmer JA, Chen M-S, Seabourn BW (2018) Registration of Avery wheat. J Plant Reg. https://doi.org/10.3198/jpr2017.11.0080crc
Harvey TL, Livers RW (1975) Resistance to wheat curl mite, Aceria tulipae Keifer, in rye and wheat-rye addition lines. Environ Entomol 4:523–526
Harvey TL, Martin TJ, Seifers DL (1994) Importance of plant resistance to insect and mite vectors in controlling virus diseases of plants: resistance to the wheat curl mite (Acari: Eriophyidae). J Agric Entomol 11:271–277
Harvey TL, Martin TJ, Seifers DL (1995) Survival of five wheat curl mites, Aceria tosichella Keifer (Acari: Eriophyidae) strains on mite resistant wheat. Exp and Appl Acarol 19:459–463
Harvey TL, Martin TJ, Seifers DL (2000) Effect of nonviruliferous wheat curl mites on yield of winter wheat. J. Agric. Urban Entomol. 17:9–13
Harvey TL, Martin TJ, Seifers DL (2002) Wheat yield reduction due to wheat curl mite (Acari: Eriophyidae) infestations. J Agric Urban Entomol 19:9–13
International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
John JA, Williams ER (1995) Cyclic and computer-generated designs, 2nd ed. St. Edmundsbury Press, Bury St. Edmunds, Suffolk, UK
Kosambi DD (1943) The estimation of map distance from recombination values. Ann Eugenic 12(3):172–175
Larson RI, Atkinson TG (1973) Wheat-Agropyron chromosome substitution lines as sources of resistance to wheat streak mosaic virus and its vector, Aceria tulipae. In: Sears ER, Sears LM (eds) Proc. 4th Int. Wheat Genetics Symposium. Columbia, Missouri, 6–11 August 1972. University of Missouri, Columbia, pp 173–177
Lazar MD, Worrall WD, Peterson GL, Fritz AK, Marshall D, Nelson LR, Rooney LW (2004) Registration of ‘TAM111’ wheat. Crop Sci 44:353–355
LGC Genomics (2017) KASP genotyping chemistry: user guide and manual. https://www.lgcgroup.com/LGCGroup/media/PDFs/Products/Genotyping/KASP-genotyping-chemistry-User-guide.pdf. Accessed 5 Jan 2018
Liu XM, Gill BS, Chen MS (2005) Hessian fly resistance gene H13 is mapped to a distal cluster of resistance genes in chromosome 6DS of wheat. Theor Appl Genet 111:243–249
Liu S, Griffey CA, Hall MD, Mckendry AL, Chen J, Brooks WS, Brown-Guedira G, Sanford D, Schmale DG (2013) Molecular characterization of field resistance to Fusarium head blight in two US soft red winter wheat cultivars. Theor Appl Genet 126:2485–2498
Liu S, Rudd JC, Bai G, Haley SD, Ibrahim AMH, Xue Q, Hays DB, Graybosch RA, Devkota RN, Amand PS (2014) Molecular markers linked to important genes in hard winter wheat. Crop Sci 54:1–18
Liu S, Assanga SO, Dhakal S, Gu X, Tan CT, Yang Y, Rudd J, Hays D, Ibrahim A, Xue Q, Chao S, Devkota R, Shachter C, Huggins T, Mohammed S, Fuentealba MP (2016) Validation of chromosomal locations of 90K array single nucleotide polymorphisms in US wheat. Crop Sci 56:364–373
Lopez-Vera EE, Nelson S, Singh RP, Basnet BR, Haley SD, Bhavani S, Huerta-Espino J, Ruiz-Medrano R, Rouse MN, Singh S (2014) Resistance to Ug99 stem rust in six bread wheat cultivars maps to chromosome arm 6DS. Theor Appl Genet 127:231–239
Malik R, Brown-Guedira GL, Smith CM, Harvey TL, Gill BS (2003) Genetic mapping of wheat curl mite resistance genes Cmc3 and Cmc4 in common wheat. Crop Sci 43:644–650
Martin TJ, Harvey TL, Bender CG, Seifers DL (1984) Control of wheat streak mosaic virus with vector resistance in wheat. Phytopathology 74:963–964
Mayer KFX, Roger J, Dolezel J, Pozniak C, Eversole K, Feuillet C, Gill B et al (2014) A chromosome-based draft sequence of the hexaploidy bread wheat (Triticum aestivum) genome. Science 345:1251788
Olson E, Rouse M, Pumphrey M, Bowden R, Gill BS, Poland J (2013) Introgression of stem rust resistance genes SrTA10187 and SrTA10171 from Aegilops tauschii to wheat. Theor Appl Genet 126:2477–2484
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7(2):e32253
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Reed E, Nunez S, Kulp D, Qian J, Reilly MP, Foulkes AS (2015) A guide to genome-wide association analysis and post-analytic interrogation. Statist Med 34:3769–3792
Ribaut JM, Banziger M, Betran J, Jiang C, Edmeades GO, Dreher K, Hoisington D (2002) Use of molecular markers in plant breeding: drought tolerance improvement in tropical maize. In: Kang MS (ed) Quantitative genetics, genomics, and plant breeding. CABI Publishing, Wallingford, UK, pp 85–100
Rosa JE, Bonnecarrere V, Perez de Vida F (2014) One-step, codominant detection of imidazolinone resistance mutations in weedy rice (Oryza sativa L.). Electron J Biotechnol 17:95–101
Rozen S, Skaletsky HJ (1998) Primer3: WWW primer tool. http://biotools.umassmed.edu/bioapps/primer3_www.cgi. Accessed 5 Jan 2018
Rudd JC, Devkota RN, Baker JA, Peterson GL, Lazar M, Bean B, Worrall D, Baughman T, Marshall D, Sutton R, Rooney LW, Nelson LR, Fritz AK, Weing Y, Morgan G, Seabourn B (2014) Registration of TAM 112 wheat. J Plant Reg 8:291–297
Schlegel R, Kynast R (1987) Conformation of 1A/1R wheat-rye chromosome translocation in the wheat variety ‘Amigo’. Plant Breed 98:57–60
Sebesta EE, Wood EA Jr, Porter DR, Webster JA, Smith EL (1995) Registration of Amigo wheat germplasm resistant to greenbug. Crop Sci 35:293
Semagn K, Babu R, Hearne S, Olsen M (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14
Slykhuis JT (1955) Aceria tulipae Keifer (Acarina: Eriophidae) in relation to spread of wheat streak mosaic. Phytopathology 45:116–128
Stewart CNJ, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14:748–751
Tan C-T, Assanga A, Zhang G, Rudd JC, Haley SD, Xue Q, Ibrahim A, Bai G, Zhang X, Byrne P, Fuentealba MP, Liu S (2017a) Development and validation of KASP markers for wheat streak mosaic virus resistance gene Wsm2. Crop Sci 57:340–349
Tan C-T, Yu H, Yang Y, Xu X, Chen M, Rudd JC, Xue Q, Ibrahim AMH, Garza L, Wang S, Sorrells ME, Liu S (2017b) Development and validation of KASP markers for the greenbug resistance gene Gb7 and the Hessian fly resistance gene H32 in wheat. Theor Appl Genet 130:1867–1884
Thomas JB, Conner RL (1986) Resistance to colonization by the wheat curl mite in Aegilops squarrosa and its inheritance after transfer to common wheat. Crop Sci 26:527–530
Thomas JB, Conner RL, Graf RJ (2004) Comparison of different sources of vector resistance for controlling wheat streak mosaic in winter wheat. Crop Sci 44:125–130
Thomson MJ, Singh N, Dwiyanti MS, Wang DR, Wright MH, Perez FA, DeClerck G, Chin JH, Malitic-Layaoen GA, Juanillas VM, Dilla-Ermita CJ, Mauleon R, Kretzschmar T, McCouch SR (2017) Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications. Rice 10:40
Turner SD (2014) qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv. https://www.r-project.org/nosvn/pandoc/qqman.html. Accessed 5 Jan 2018
Van Ooijen JW (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93:343–349
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploidy wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol 12:787–796
Wang H, Zhang H, Du R, Chen G, Liu B, Yang Y, Qin L, Cheng E et al (2016) Identification and validation of QTLs controlling multiple traits in sorghum. Crop Pasture Sci 67:193–203
Whelan EDP, Hart GE (1988) A spontaneous translocation that transfers wheat curl mite resistance from decaploid Agropyron elongatum to common wheat. Genome 31:289–292
Whelan EDP, Thomas JB (1989) Chromosomal location in common wheat of a gene (Cmc1) from Aegilops squarrosa that conditions resistance to colonization by the wheat curl mite. Genome 32:1033–1036
Wilkinson PA, Winfield MO, Barker GLA, Allen AM, Burridge A, Coghill JA, Burridge A, Edwards KJ (2012) CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinf 13:219. http://www.cerealsdb.uk.net//cerealgenomics/CerealsDB/blast_WGS.php. Accessed 24 Jan 2018
Williams E, Piepho HP, Whitaker D (2011) Augmented p-rep designs. Biom J 53:19–27. https://doi.org/10.1002/bimj.201000102
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Zhang G, Martin TJ, Fritz AK, Miller R, Chen M-S, Bowden RL, Bai G (2016) Registration of ‘Joe’ hard white winter wheat. J. Plant Reg. 10:283–286
Zhou H, Liu S, Liu Y, You J, Deng M, Ma J, Chen G et al (2016) Mapping and validation of major quantitative trait loci for kernel length in wild barley (Hordeum vulgare ssp. spontaneum). BMC Genet 17:130
Acknowledgments
The authors acknowledge Jason Baker at Texas A&M AgriLife Research Center in Amarillo for technical help and development of the population. We acknowledge Dr. Chenggen Chu and Dev R. Poudel for their critical review and suggestions. We also thank Dr. Charlie Rush for providing access to the ABI 7500 instrument for running KASP assays.
Author contribution statement
S. Dhakal conducted phenotyping experiments for WCM, performed all aspects of data analysis and wrote the manuscript; C.-T. Tan confirmed KASP markers and ran the analysis; H. Yu performed KASP assays and rephenotyped some heterogeneous lines; L. Garza prepared the WCM experiments; J.C. Rudd, Q. Xue and A.M.H. Ibrahim provided overall support and help with data interpretation; R.N. Devkota coordinated advancement of the mapping population; V. Anderson and S.D. Haley conducted the GWAS for the breeding nursery lines, analyzed related data, interpreted the results, and provided editorial input on the draft manuscript; S. Liu designed the experiments and provided suggestions for data analysis, interpretation and writing of the manuscript.
Funding
This research was partially supported by the Texas Wheat Producers Board, Texas A&M AgriLife Research and the National Research Initiative Competitive Grants 2017-67007-25939 from the USDA National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The experiments comply with the ethical standards in the country in which they were performed.
Rights and permissions
About this article
Cite this article
Dhakal, S., Tan, CT., Anderson, V. et al. Mapping and KASP marker development for wheat curl mite resistance in “TAM 112” wheat using linkage and association analysis. Mol Breeding 38, 119 (2018). https://doi.org/10.1007/s11032-018-0879-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0879-x