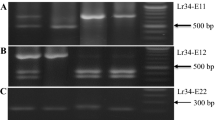
Abstract
As introns are vulnerable to changes such as insertions and deletions when exposed to various evolutionary forces, they constitute a repository for developing genetic markers based on intron length polymorphisms (ILP). This study developed a set of genetic markers that use the potential intron length polymorphism in resistance gene analogs (RGAs) in Zea mays. By searching the genome of Zea mays B73 for the homologs of 73 R genes which have already been identified in plants, we found 861 RGAs, 632 of which have at least one intron that can serve as putative markers targeting the intron length polymorphism in RGAs (RGA-ILP). We developed 1972 candidate markers via electronic PCR (e-PCR) with primer pairs designed in each pair of exonic regions that flank an intron. Furthermore, the performance of RGA-ILP among four maize inbred lines (Huangzao4, B73, Mo17, and Dan340) was evaluated with 69 pairs of randomly selected primers. Of them, 46.4% showed bands that had discriminating length polymorphism, and between any two of the inbred lines the proportion of polymorphism ranged from 23.2 to 31.9%. To make it convenient to use these markers for those interested in molecular breeding of disease-resistant maize, we provide all related information in a web-based database named MaizeRGA, which is available at http://www.sicau.edu.cn/web/yms/rga/maizeRGA.html.


Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Ayliffe MA, Lagudah ES (2004) Molecular genetics of disease resistance in cereals. Ann Bot 94:765–773
Balint-Kurti PL, Johal GS (2009) Maize disease resistance. In: Bennetzen JL, Hake SC (eds) Handbook of maize: its biology. Springer, New York, pp 229–250
Bentolila S, Guitton C, Bouvet N, Sailland A, Nykaza S, Freyssinet G (1991) Identification of an RFLP marker tightly linked to the Ht1 gene in maize. Theor Appl Genet 82:393–398
Chung CL, Jamann T, Longfello J, Nelson R (2010) Characterization and fine-mapping of a resistance locus for northern leaf blight in maize bin 8.06. Theor Appl Genet 121:205–227
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. MPMI 10:968–978
Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Tony P (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11:1365–1376
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Dangl JL, Jones JDG (2006) The plant immune system. Nature 444:323–329
Delorenzi M, Speed T (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18:617–625
Duble CM, Melchinger AE, Kuntze L, Stork A, Lübberstedt T (2000) Molecular mapping and gene action of Scm1 and Scm2, two major QTL contributing to SCMV resistance in maize. Plant Breed 119:299–303
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Edwards MD, Helentjaris T, Wright S, Stuber CW (1992) Molecular-marker-facilitated investigations of quantitative trait loci in maize. Theor Appl Genet 83:765–774
Fan XM, Tan J, Zhang SH, Li MS, Huang YX, Yang JY, Peng ZB, Li XH (2003) Heterotic grouping for 25 tropical maize inbreds and 4 temperate maize inbreds by SSR markers. Acta Agron Sin 29:835–840
Feltus FA, Singh HP, Lohithaswa HC, Schulze SR, Silva TD, Paterson AH (2006) A comparative genomics strategy for targeted discovery of single-nucleotide polymorphisms and conserved-noncoding sequences in orphan crops. Plant Physiol 140:1183–1191
Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A (2008) The Pfam protein families database. Nucl Acids Res 36:D281–D288
Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8:967–974
Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, Ware DH, Buckler ES (2009) A first-generation haplotype map of maize. Science 326:1115–1117
Gou MY, Su N, Zheng J, Huai JL, Wu GH, Zhao JF, He JG, Tang DZ, Yang SH, Wang GY (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60:757–770
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Biol 48:575–607
Ingvardsen CR, Xing YZ, Frei UK, Lübberstedt T (2010) Genetic and physical fine mapping of Scmv2, a potyvirus resistance gene in maize. Theor Appl Genet 120:1621–1634
Jiang L, Ingvardsen CR, Lübberstedt T, Xu ML (2008) The Pic19 NBS-LRR gene family members are closely linked to Scmv1, but not involved in maize resistance to sugarcane mosaic virus. Genome 51:673–684
Jones JDG (2001) Putting knowledge of plant disease resistance genes to work. Curr Opin Plant Biol 4:281–287
Marsan PA, Gorni C, Chitto A, Redaelli R, Vijk RV, Stam P, Motto M (2001) Identification of QTLs for grain yield and grain-related traits of maize (Zea mays L.) using an AFLP map, different testers, and cofactor analysis. Theor Appl Genet 102:230–243
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8:129–134
Ming R, Brewbaker JL, Pratt RC, Musket TA, McMullen MD (1997) Molecular mapping of a major gene conferring resistance to maize mosaic virus. Theor Appl Genet 95:271–275
Moller S, Croning MDR, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653
Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Pflieger S, Lefebvre V, Causse M (2001) The candidate gene approach in plant genetics: a review. Mol Breed 7:275–291
Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14:21–29
Quint M, Mihaljevic R, Dussle CM, Xu ML, Melchinger AE, Lübberstedt T (2002) Development of RGA-CAPS markers and genetic mapping of candidate genes for sugarcane mosaic virus resistance in maize. Theor Appl Genet 105:355–363
Ramakrishna W, Emberton J, SanMiguel P, Ogden M, Llaca V, Messing J, Bennetzen JL (2002) Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol 130:1728–1738
Ribaut JM, Betran J (1999) Single large-scale marker-assisted selection (SLS-MAS). Mol Breed 5:531–541
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sanseverino W, Roma G, Simone MD, Faino L, Melito S, Stupka E, Frusciante L, Ercolano MR (2009) PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucl Acids Res 38:D814–D821
Sanz-Alferez S, Richter TE, Hulbert SH, Bennetzen JL (1995) The Rp3 disease resistance gene of maize: mapping and characterization of introgressed alleles. Theor Appl Genet 91:25–32
Schnable PS, Ware D, Fulton RS et al (2009) The B73 Maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Schuler GD (1997) Sequence mapping by electronic PCR. Genome Res 7:541–550
Shang W, Zhou RH, Jia JZ, Gao LF (2010) RGA-ILP, a new type of functional molecular markers in bread wheat. Euphytica 172:263–273
Wang XS, Wu WR, Jin GL, Zhu J (2005a) Genome-wide identification of R genes and exploitation of candidate RGA markers in rice. Chin Sci Bull 50:1120–1125
Wang XS, Zhao XQ, Zhu J, Wu WR (2005b) Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Res 12:417–427
Wei H, Fu Y, Arora R (2005) Intron-flanking EST-PCR markers: from genetic marker development to gene structure analysis in Rhododendron. Theor Appl Genet 111:1347–1356
Welz HG, Schechert A, Pernet A, Pixley KV, Geiger HH (1998) A gene for resistance to the maize streak virus in the African CIMMYT maize inbred line CML202. Mol Breed 4:147–154
Wisser RJ, Balint-Kurti PJ, Nelson RJ (2006) The genetic architecture of disease resistance in maize: a synthesis of published studies. Phytopathology 96:120–129
Xiao WK, Xu ML, Zhao JR, Wang FG, Li JS, Dai JR (2006) Genome-wide isolation of resistance gene analogs in maize (Zea mays L.). Theor Appl Genet 113:63–72
Xiao WK, Zhao J, Fan SC, Li L, Dai JR, Xu ML (2007) Mapping of genome-wide resistance gene analogs (RGAs) in maize (Zea mays L.). Theor Appl Genet 115:501–508
Xu ML, Melchinger AE, Xia XC, Lübberstedt T (1999) High-resolution mapping of loci conferring resistance to sugarcane mosaic virus in maize using RFLP, SSR, and AFLP markrs. Mol Gen Genet 261:574–581
Xu ML, Melchinger AE, Lübberstedt T (2000) Origin of Scm1 and Scm2-two loci conferring resistance to sugarcane mosaic virus (SCMV) in maize. Theor Appl Genet 100:934–941
Yang L, Jin GL, Zhao XQ, Zheng Y, Xu ZH, Wu WR (2007) PIP: a database of potential intron polymorphism markers. Bioinformatics 23:2174–2177
Zhang LQ, Peek AS, Dunams D, Gaut BS (2002) Population genetics of duplicated disease-defense genes, hm1 and hm2, in maize (Zea mays ssp. mays L.) and its wild ancestor (Zea mays ssp. parviglumis). Genetics 162:851–860
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Gen Genet 271:402–415
Acknowledgments
We thank Dr. Chen Guoyue from the Triticeae Research Institute of Sichuan Agricultural University for helpful discussions during this study. The authors thank two anonymous reviewers and Dr Yann Klimentidis for critical reading. This work is supported by the grants from National Natural Science Foundation of China (No.30800687 and 31071434), Applied Basic Research Program of Sichuan Provincial Science and Technology Department (No.2008JY0096), and the ‘863’ High Technology Development Program (No.2009AA10AA03_2).
Author information
Authors and Affiliations
Corresponding authors
Additional information
H. Liu and Y. Lin have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, H., Lin, Y., Chen, G. et al. Genome-scale identification of resistance gene analogs and the development of their intron length polymorphism markers in maize. Mol Breeding 29, 437–447 (2012). https://doi.org/10.1007/s11032-011-9560-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-011-9560-3