Abstract
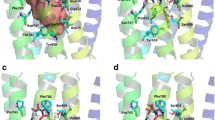
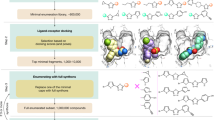
A glutaminyl cyclase (QC) fragment library was in silico selected by disconnection of the structure of known QC inhibitors and by lead-like 2D virtual screening of the same set. The resulting fragment library (204 compounds) was acquired from commercial suppliers and pre-screened by differential scanning fluorimetry followed by functional in vitro assays. In this way, 10 fragment hits were identified (\(\sim \)5 % hit rate, best inhibitory activity: 16 \(\upmu \hbox {M}\)). The in vitro hits were then docked to the active site of QC, and the best scoring compounds were analyzed for binding interactions. Two fragments bound to different regions in a complementary manner, and thus, linking those fragments offered a rational strategy to generate novel QC inhibitors. Based on the structure of the virtual linked fragment, a 77-membered QC target focused library was selected from vendor databases and docked to the active site of QC. A PubChem search confirmed that the best scoring analogues are novel, potential QC inhibitors.










Similar content being viewed by others
References
Nadin A, Hattotuwagama C, Churcher I (2012) Lead-oriented synthesis: a new opportunity for synthetic chemistry. Angew Chem Int Ed 51:1114–1122. doi:10.1002/anie.201105840
Hann MM, Keserü GM (2012) Finding the sweet spot: the role of nature and nurture in medicinal chemistry. Nat Rev Drug Discov 11:355–365. doi:10.1038/nrd3701
Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P (2012) Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov 11:873–886. doi:10.1038/nrd3847
Baker M (2013) Fragment-based lead discovery grows up. Nat Rev Drug Discov 12:5–7. doi:10.1038/nrd3926
Sanders WJ, Nienaber VL, Lerner CG, McCall JO, Merrick SM, Swanson SJ, Harlan JE, Stoll VS, Stamper GF, Betz SF, Condroski KR (2004) Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization. J Med Chem 47:1709–1718. doi:10.1021/jm049575n
Gill AL, Frederickson M, Cleasby A, Woodhead SJ, Carr MG, Woodhead AJ, Walker MT, Congreve MS, Devine LA, Tisi D, O’Reilly M (2005) Identification of novel p38\(\alpha \) MAP kinase inhibitors using fragment-based lead generation. J Med Chem 48:414–426. doi:10.1021/jm030497y
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. doi:10.1016/S0169-409X(00)00129-0
Congreve M, Carr R, Murray C, Jhoti H (2003) A rule of three for fragment-based lead discovery? Drug Discov Today 8:876–877. doi:10.1016/S1359-6446(03)02831-9
Jhoti H, Williams G, Rees DC, Murray CW (2013) The rule of three for fragment-based drug discovery: where are we now? Nat Rev Drug Discov 12:644–645. doi:10.1038/nrd3926-c1
Scott DE, Coyne AG, Hudson SA, Abell C (2012) Fragment-based approaches in drug discovery and chemical biology. Biochemistry 51:4990–5003. doi:10.1021/bi3005126
Murray CW, Rees DC (2009) The rise of fragment-based drug discovery. Nat Chem 1:187–192. doi:10.1038/nchem.217
Kranz JK, Schalk-Hihi C (2011) Protein thermal shifts to identify low molecular weight fragments. Methods Enzymol 493:277–298. doi:10.1016/B978-0-12-381274-2.00011-X
Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi:10.1038/nprot.2007.321
Cynis H, Scheel E, Saido TC, Schilling S, Demuth H-U (2008) Amyloidogenic processing of amyloid precursor protein: evidence of a pivotal role of glutaminyl cyclase in generation of pyroglutamate-modified amyloid-\(\beta \). Biochemistry 47:7405–7413. doi:10.1021/bi800250p
Nussbaum JM, Schilling S, Cynis H, Silva A, Swanson E, Wangsanut T, Tayler K, Wiltgen B, Hatami A, Rönicke R, Reymann K (2012) Prion-like behaviour and tau-dependent cytotoxicity of pyroglutamylated amyloid-[bgr]. Nature 485:651–655. doi:10.1038/nature11060
Schilling S, Zeitschel U, Hoffmann T, Heiser U, Francke M, Kehlen A, Holzer M, Hutter-Paier B, Prokesch M, Windisch M, Jagla W, Schlenzig D, Lindner C, Rudolph T, Reuter G, Cynis H, Montag D, Demuth HU, Rossner S (2008) Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer’s disease-like pathology. Nat Med 14:1106–1111. doi:10.1038/nm.1872
Schilling S, Appl T, Hoffmann T, Cynis H, Schulz K, Jagla W, Friedrich D, Wermann M, Buchholz M, Heiser U, Von Hörsten S (2008) Inhibition of glutaminyl cyclase prevents pGlu-\(\text{ A }\beta \) formation after intracortical/hippocampal microinjection in vivo/in situ. J Neurochem 106:1225–1236. doi:10.1111/j.1471-4159.2008.05471.x
Jimenez-Sanchez M, Lam W, Hannus M, Sönnichsen B, Imarisio S, Fleming A, Tarditi A, Menzies F, Dami TE, Xu C, Gonzalez-Couto E (2015) siRNA screen identifies QPCT as a druggable target for Huntington’s disease. Nat Chem Biol 11:347–354. doi:10.1038/nchembio.1790
Ramsbeck D, Buchholz M, Koch B, Bohme L, Hoffmann T, Demuth HU, Heiser U (2013) Structure-activity relationships of benzimidazole-based glutaminyl cyclase inhibitors featuring a heteroaryl scaffold. J Med Chem 56:6613–6625. doi:10.1021/jm4001709
Schilling S, Niestroj AJ, Rahfeld JU, Hoffmann T, Wermann M, Zunkel K, Wasternack C, Demuth HU (2003) Identification of human glutaminyl cyclase as a metalloenzyme. Potent inhibition by imidazole derivatives and heterocyclic chelators. J Biol Chem 278:49773–49779. doi:10.1074/jbc.M309077200
Buchholz M, Heiser U, Schilling S, Niestroj AJ, Zunkel K, Demuth H-U (2006) The first potent inhibitors for human glutaminyl cyclase: synthesis and structure-activity relationship. J Med Chem 49:664–677. doi:10.1021/jm050756e
Buchholz M, Hamann A, Aust S, Brandt W, Bohme L, Hoffmann T, Schilling S, Demuth HU, Heiser U (2009) Inhibitors for human glutaminyl cyclase by structure based design and bioisosteric replacement. J Med Chem 52:7069–7080. doi:10.1021/jm900969p
Tran PT, Hoang VH, Thorat SA, Kim SE, Ann J, Chang YJ, Nam DW, Song H, Mook-Jung I, Lee J, Lee J (2013) Structure-activity relationship of human glutaminyl cyclase inhibitors having an N-(5-methyl-1H-imidazol-1-yl)propyl thiourea template. Bioorg Med Chem 21:3821–3830. doi:10.1016/j.bmc.2013.04.005
Origin 8.0, OriginLab, Northampton, MA, USA (2007)
Schilling S, Hoffmann T, Wermann M, Heiser U, Wasternack C, Demuth H-U (2002) Continuous spectrometric assays for glutaminyl cyclase activity. Anal Biochem 303:49–56. doi:10.1006/abio.2001.5560
Tounge BA, Parker MH (2011) Designing a diverse high-quality library for crystallography-based FBDD screening. Methods Enzymol 493:3–20. doi:10.1016/B978-0-12-381274-2.00001-7
Tovar A, Eckert H, Bajorath J (2007) Comparison of 2D fingerprint methods for multiple-template similarity searching on compound activity classes of increasing structural diversity. ChemMedChem 2:208–217. doi:10.1002/cmdc.200600225
Willett P, Winterman V (1986) A comparison of some measures for the determination of inter-molecular structural similarity. Quant Struct Act Relatsh 5:18–25. doi:10.1002/qsar.19860050105
Instant JChem 6.4, JChem for Excel 14.8, ChemAxon Ltd., Budapest, Hungary (2014)
Similarity Manager 1.0, CompuDrug Int., Sedona, AZ, USA (2006)
Bioster 4.1, Digital Chemistry Ltd, Sheffield, UK (2008)
Ruiz-Carrillo D, Koch B, Parthier C, Wermann M, Dambe T, Buchholz M, Ludwig H-H, Heiser U, Rahfeld J-U, Stubbs MT, Schilling S (2011) Structures of glycosylated mammalian glutaminyl cyclases reveal conformational variability near the active center. Biochemistry 50:6280–6288. doi:10.1021/bi200249h
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. doi:10.1093/nar/28.1.235
Small-Molecule Drug Discovery Suite 2015-3 (2015) Schrödinger Suite 2015-3 Induced Fit Docking protocol; Glide version 6.8, Schrödinger, LLC, New York, NY, 2015; Prime version 4.1, Schrödinger, LLC, New York, NY
Sherman W, Day T, Jacobson MP, Friesner RA, Farid R (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553. doi:10.1021/jm050540c
Steinbrecher TB, Dahlgren M, Cappel D, Lin T, Wang L, Krilov G, Abel R, Friesner R, Sherman W (2015) Accurate binding free energy predictions in fragment optimization. J Chem Inf Model 55:2411–2420. doi:10.1021/acs.jcim.5b00538
Schultes S, de Graaf C, Haaksma EE, de Esch IJ, Leurs R, Krämer O (2010) Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discov Today Technol 7:e157–e162. doi:10.1016/j.ddtec.2010.11.003
Ferenczy GG, Keseru GM (2015) The impact of binding thermodynamics on medicinal chemistry optimizations. Future Med Chem 7:1285–1303. doi:10.4155/fmc.15.63
Acknowledgements
This work was partially supported by OTKA grants NK100482 and K115698 (to C. M. and I. S.), by the Richter Thematic Research Grant (to M. S., Z. L, S. C., G. D) and by the Hungarian Government (National Research, Technology and Innovation Fund, KMR_12-1-2012-0218, to M. S., Z. L, S. C., G. D). We are grateful to ChemAxon Ltd., Budapest for providing Instant JChem software for 2D similarity search and physicochemical parameter prediction.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Szaszkó, M., Hajdú, I., Flachner, B. et al. Identification of potential glutaminyl cyclase inhibitors from lead-like libraries by in silico and in vitro fragment-based screening. Mol Divers 21, 175–186 (2017). https://doi.org/10.1007/s11030-016-9717-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-016-9717-4