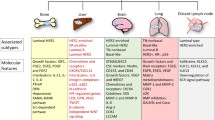
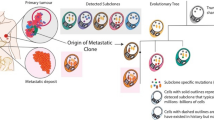
Abstract
Breast cancer is the most commonly diagnosed and the second highest cause of cancer-related mortality. Although major breakthroughs have emerged during the past decades concerning the characterization of major malignant tumors hallmarks, little is known about the molecular process that sustains the most deadly feature of cancer: metastasis to distant organs. In fact, this colonization of tumor cells to secondary sites is not random but rather orientated, and depends on several signalling events that are not fully elucidated yet. Understanding the precise molecular and cellular mechanisms accountable for the specific invasion of tissues by breast cancer cells is likely to be important for developing new therapeutic strategies to effectively prevent metastasis in patients diagnosed with early cancer lesions. Here, we briefly describe a multidisciplinary approach based on the molecular profiling of breast cancer metastases, the elaboration of prognostic gene signatures, the clinical validation and the experimental confirmation using cell and animal models to better address breast cancer metastasis. This methodology can be considered as a useful workflow to identify and validate the genes that trigger and support organ tropism of breast cancer cells during metastasis.


Similar content being viewed by others
Abbreviations
- IHC:
-
immunohistochemistry
- EMT:
-
epithelial mesenchymal transition
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi:10.1016/j.cell.2011.02.013.
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8. doi:10.1038/nrc1098.
Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–72. doi:10.1038/nrc865.
Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8(2):98–101.
Lee YT. Breast carcinoma: pattern of metastasis at autopsy. J Surg Oncol. 1983;23(3):175–80.
Patanaphan V, Salazar OM, Risco R. Breast cancer: metastatic patterns and their prognosis. South Med J. 1988;81(9):1109–12.
Weigelt B, Peterse JL, Van’t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi:10.1038/nrc1670.
Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–84. doi:10.1038/nrc2622.
Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, et al. Metastatic patterns in adenocarcinoma. Cancer. 2006;106(7):1624–33. doi:10.1002/cncr.21778.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi:10.1056/NEJMoa021967.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi:10.1056/NEJMoa041588.
Driouch K, Landemaine T, Sin S, Wang S, Lidereau R. Gene arrays for diagnosis, prognosis and treatment of breast cancer metastasis. Clin Exp Metastasis. 2007;24(8):575–85. doi:10.1007/s10585-007-9110-x.
Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11(10):735–48. doi:10.1038/nrc3125.
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi:10.1038/nature08989.
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi:10.1126/science.1096502.
Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–13. doi:10.1038/nature09460.
Hunter K, Welch DR, Liu ET. Genetic background is an important determinant of metastatic potential. Nat Genet. 2003;34(1):23–4. doi:10.1038/ng0503-23b. author reply 5.
Hunter KW, Alsarraj J. Gene expression profiles and breast cancer metastasis: a genetic perspective. Clin Exp Metastasis. 2009;26(6):497–503. doi:10.1007/s10585-009-9249-8.
Dutertre M, Lacroix-Triki M, Driouch K, de la Grange P, Gratadou L, Beck S, et al. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res. 2010;70(3):896–905. doi:10.1158/0008-5472.CAN-09-2703.
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. doi:10.1038/nature08514.
Chen X, Jorgenson E, Cheung ST. New tools for functional genomic analysis. Drug Discov Today. 2009;14(15–16):754–60. doi:10.1016/j.drudis.2009.05.005.
Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet. 2011;12(7):487–98. doi:10.1038/nrg3013.
Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi:10.1016/j.ccr.2007.08.030.
Gobeil S, Zhu X, Doillon CJ, Green MR. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 2008;22(21):2932–40. doi:10.1101/gad.1714608.
Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–12. doi:10.1038/nature08460.
Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–72. doi:10.1038/nature09627.
Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348–53. doi:10.1126/science.1212728.
Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16(4):324–35. doi:10.1016/j.ccr.2009.08.015.
Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145(1):145–58. doi:10.1016/j.cell.2011.03.012.
Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, et al. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11(11):1297–304. doi:10.1038/ncb1974.
Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–50. doi:10.1038/nature10350.
Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2(6):418–27. doi:10.1038/35076576.
Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25(6):534–55. doi:10.1101/gad.2017311.
Landemaine T, Jackson A, Bellahcene A, Rucci N, Sin S, Abad BM, et al. A six-gene signature predicting breast cancer lung metastasis. Cancer Res. 2008;68(15):6092–9. doi:10.1158/0008-5472.CAN-08-0436.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi:10.1073/pnas.191367098.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi:10.1038/35021093.
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi:10.1073/pnas.0932692100.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7. doi:7911.
Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–49.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–24. doi:10.1038/nature03799.
Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–9. doi:10.1038/nature08021.
Sanz-Pamplona R, Aragues R, Driouch K, Martin B, Oliva B, Gil M, et al. Expression of endoplasmic reticulum stress proteins is a candidate marker of brain metastasis in both ErbB-2+ and ErbB-2- primary breast tumors. Am J Pathol. 2011;179(2):564–79. doi:10.1016/j.ajpath.2011.04.037.
Sin S, Bonin F, Petit V, Meseure D, Lallemand F, Bieche I, et al. Role of the focal adhesion protein kindlin-1 in breast cancer growth and lung metastasis. J Natl Cancer Inst. 2011;103(17):1323–37. doi:10.1093/jnci/djr290.
West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98(20):11462–7. doi:10.1073/pnas.201162998.
Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–24. doi:10.1038/nm733.
Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102(39):13909–14. doi:10.1073/pnas.0506517102.
Eckhardt BL, Parker BS, van Laar RK, Restall CM, Natoli AL, Tavaria MD, et al. Genomic analysis of a spontaneous model of breast cancer metastasis to bone reveals a role for the extracellular matrix. Mol Cancer Res. 2005;3(1):1–13.
Lorger M, Lee H, Forsyth JS, Felding-Habermann B. Comparison of in vitro and in vivo approaches to studying brain colonization by breast cancer cells. J Neurooncol. 2011;104(3):689–96. doi:10.1007/s11060-011-0550-4.
Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15(1):87–96. doi:10.1016/j.gde.2004.12.002.
Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–54. doi:10.1038/nrc822.
Moreno-Bueno G, Peinado H, Molina P, Olmeda D, Cubillo E, Santos V, et al. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc. 2009;4(11):1591–613. doi:10.1038/nprot.2009.152.
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–9. doi:10.1038/nprot.2006.339.
Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3(5):362–74. doi:10.1038/nrc1075.
Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12(2–3):153–62. doi:10.1007/s10911-007-9047-3.
Kim IS, Baek SH. Mouse models for breast cancer metastasis. Biochem Biophys Res Commun. 2010;394(3):443–7. doi:10.1016/j.bbrc.2010.03.070.
Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8(4):212. doi:10.1186/bcr1530.
Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27(2):83–90. doi:10.1007/s10585-010-9305-4.
Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–66. doi:10.1016/j.ccr.2009.07.016.
Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–8006. doi:10.1158/0008-5472.CAN-05-4399.
Cailleau R, Olive M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14(11):911–5.
Kuperwasser C, Dessain S, Bierbaum BE, Garnet D, Sperandio K, Gauvin GP, et al. A mouse model of human breast cancer metastasis to human bone. Cancer Res. 2005;65(14):6130–8. doi:10.1158/0008-5472.CAN-04-1408.
Yoneda T, Sasaki A, Mundy GR. Osteolytic bone metastasis in breast cancer. Breast Cancer Res Treat. 1994;32(1):73–84.
Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res. 2001;16(8):1486–95. doi:10.1359/jbmr.2001.16.8.1486.
Munoz R, Man S, Shaked Y, Lee CR, Wong J, Francia G, et al. Highly efficacious nontoxic preclinical treatment for advanced metastatic breast cancer using combination oral UFT-cyclophosphamide metronomic chemotherapy. Cancer Res. 2006;66(7):3386–91. doi:10.1158/0008-5472.CAN-05-4411.
Bonnomet A, Syne L, Brysse A, Feyereisen E, Thompson EW, Noel A, et al. A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene. 2011. doi:10.1038/onc.2011.540.
Vantyghem SA, Allan AL, Postenka CO, Al-Katib W, Keeney M, Tuck AB, et al. A new model for lymphatic metastasis: development of a variant of the MDA-MB-468 human breast cancer cell line that aggressively metastasizes to lymph nodes. Clin Exp Metastasis. 2005;22(4):351–61. doi:10.1007/s10585-005-0745-1.
Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–13. doi:10.1158/0008-5472.CAN-08-2741.
Xia TS, Wang GZ, Ding Q, Liu XA, Zhou WB, Zhang YF et al. Bone metastasis in a novel breast cancer mouse model containing human breast and human bone. Breast Cancer Res Treat. 2011. doi:10.1007/s10549-011-1496-0
Khalili P, Arakelian A, Chen G, Singh G, Rabbani SA. Effect of Herceptin on the development and progression of skeletal metastases in a xenograft model of human breast cancer. Oncogene. 2005;24(44):6657–66. doi:10.1038/sj.onc.1208790.
Harrell JC, Dye WW, Allred DC, Jedlicka P, Spoelstra NS, Sartorius CA, et al. Estrogen receptor positive breast cancer metastasis: altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer Res. 2006;66(18):9308–15. doi:10.1158/0008-5472.CAN-06-1769.
Rucci N, Ricevuto E, Ficorella C, Longo M, Perez M, Di Giacinto C, et al. In vivo bone metastases, osteoclastogenic ability, and phenotypic characterization of human breast cancer cells. Bone. 2004;34(4):697–709. doi:10.1016/j.bone.2003.07.012.
Care A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, et al. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61(17):6532–9.
Couillard S, Labrie C, Gauthier S, Merand Y, Singh SM, Candas B, et al. Long-term inhibitory effect of the orally active and pure antiestrogen EM-800 on the growth of human breast cancer xenografts in nude mice. Int J Cancer. 2000;85(3):424–9. doi:10.1002/(SICI)1097-0215(20000201)85:3<424::AID-IJC20>3.0.CO;2-S.
Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–405.
Lelekakis M, Moseley JM, Martin TJ, Hards D, Williams E, Ho P, et al. A novel orthotopic model of breast cancer metastasis to bone. Clin Exp Metastasis. 1999;17(2):163–70.
Li Z, Schem C, Shi YH, Medina D, Zhang M. Increased COX2 expression enhances tumor-induced osteoclastic lesions in breast cancer bone metastasis. Clin Exp Metastasis. 2008;25(4):389–400. doi:10.1007/s10585-007-9117-3.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nola, S., Sin, S., Bonin, F. et al. A Methodological Approach to Unravel Organ-Specific Breast Cancer Metastasis. J Mammary Gland Biol Neoplasia 17, 135–145 (2012). https://doi.org/10.1007/s10911-012-9256-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-012-9256-2