Abstract
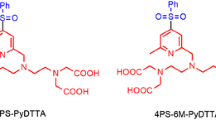
Pseudocontact shifts (PCS) generated by lanthanide chelating tags yield valuable restraints for investigating protein structures, dynamics and interactions in solution. In this work, dysprosium-, thulium- and terbium-complexes of eight-fold methylated 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid tags [DOTA-M8-(4R4S)-SSPy] are presented that induce large pseudocontact shifts up to 5.5 ppm and adopt exclusively the square antiprismatic conformation. This is in contrast to our earlier findings on complexes of the stereoisomeric DOTA-M8-(8S)-SSPy, where significant amounts of the twisted square antiprismatic conformer for the Dy tag were observed. The Dy-, Tm-, Tb- and Lu-complexes of DOTA-M8-(4R4S)-SSPy were conjugated to ubiquitin S57C and selectively 15N leucine labeled human carbonic anhydrase II S50C, resulting in only one set of signals. Furthermore, we investigated the conformation of the thulium- and dysprosium-complexes in vacuo and with implicit water solvent using density functional theory calculations. The calculated energy differences between the two different conformations (7.0–50.5 kJ/mol) and experimental evidence from the corresponding ytterbium- and yttrium-complexes clearly suggest a SAP [Λ(δδδδ)] geometry for the complexes presented in this study. The lanthanide chelating tag studied in this work offer insights into the solution structure of proteins by inducing strong pseudocontact shifts, show different tensor properties compared to its predecessor, enables a convenient assignment procedure, is accessed by a more economic synthesis than its predecessor and constitutes a highly promising starting point for further developments of lanthanide chelating tags.







Similar content being viewed by others
References
Avvaru BS, Kim CU, Sippel KH, Gruner SM, Agbandje-McKenna M, Silverman DN, McKenna R (2010) A short, strong hydrogen bond in the active site of human carbonic anhydrase II. Biochemistry 49:249–251
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38:3098–3100
Benetollo F, Bombieri G, Calabi L, Aime S, Botta M (2003) Structural variations across the lanthanide series of macrocyclic DOTA complexes: insights into the design of contrast agents for magnetic resonance imaging. Inorg Chem 42:148–157
Blahut J, Hermann P, Tosner Z, Platas-Iglesias C (2017) A combined NMR and DFT study of conformational dynamics in lanthanide complexes of macrocyclic DOTA-like ligands. PCCP 19:26662–26671
Bleaney B (1972) Nuclear magnetic resonance shifts in solution due to lanthanide ions. J Magn Reson 8:91–100
Brewer KD, Bacaj T, Cavalli A, Camilloni C, Swarbrick JD, Liu J, Zhou A, Zhou P, Barlow N, Xu J, Seven AB, Prinslow EA, Voleti R, Häussinger D, Bonvin AM, Tomchick DR, Vendruscolo M, Graham B, Südhof TC, Rizo J (2015) Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution. Nat Struct Mol Biol 22:555–564
Chen JL, Zhao Y, Gong YJ, Pan B-B, Wang X, Su XC (2018) Stable and rigid DTPA-like paramagnetic tags suitable for in vitro and in situ protein NMR analysis. J Biomol NMR 70:77–92
Cosentino U, Villa A, Pitea D, Moro G, Barone V, Maiocchi A (2002) Conformational characterization of lanthanide(III)-DOTA complexes by ab initio investigation in vacuo and in aqueous solution. J Am Chem Soc 124:4901–4909
Cramer CJ, Truhlar DG (2009) Density functional theory for transition metals and transition metal chemistry. PCCP 11:10757–10816
Dolg M, Stoll H, Savin A, Preuss H (1989) Energy-adjusted pseudopotentials for the rare earth elements. Theor Chim Acta 75:173–194
Dolg M, Stoll H, Preuss H (1993) A combination of quasirelativistic pseudopotential and ligand field calculations for lanthanoid compounds. Theor Chim Acta 85:441–450
Funk AM, Finney K, Harvey P, Kenwright AM, Neil ER, Rogers NJ, Kanthi Senanayake P, Parker D (2015) Critical analysis of the limitations of Bleaney’s theory of magnetic anisotropy in paramagnetic lanthanide coordination complexes. Chem Sci 6:1655–1662
Gaponenko V, Altieri AS, Li J, Byrd RA (2002) Breaking symmetry in the structure determination of (large) symmetric protein dimers. J Biomol NMR 24:143–148
Gempf KL, Butler SJ, Funk AM, Parker D (2013) Direct and selective tagging of cysteine residues in peptides and proteins with 4-nitropyridyl lanthanide complexes. Chem Commun 49:9104–9106
Graham B, Loh CT, Swarbrick JD, Ung P, Shin J, Yagi H, Jia X, Chhabra S, Barlow N, Pintacuda G, Huber T, Otting G (2011) DOTA-amide lanthanide tag for reliable generation of pseudocontact shifts in protein NMR spectra. Bioconj Chem 22:2118–2125
Grimme S (2005) Accurate calculation of the heats of formation for large main group compounds with spin-component scaled MP2 methods. J Phys Chem A 109:3067–3077
Grimmel S, Schoendorff G, Wilson AK (2016) Gauging the performance of density functionals for lanthanide-containing molecules. J Chem Theory Comput 12:1259–1266
Häussinger D, Huang JR, Grzesiek S (2009) DOTA-M8: an extremely rigid, high-affinity lanthanide chelating tag for PCS NMR spectroscopy. J Am Chem Soc 131:14761–14767
Helm L (2006) Relaxivity in paramagnetic systems: theory and mechanisms. Prog Nucl Magn Reson Spectrosc 49:45–64
Hikone Y, Hirai G, Mishima M, Inomata K, Ikeya T, Arai S, Shirakawa M, Sodeoka M, Ito Y (2016) A new carbamidemethyl-linked lanthanoid chelating tag for PCS NMR spectroscopy of proteins in living HeLa cells. J Biomol NMR 66:99–110
Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C (2004) Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR 29:339–349
Keizers PHJ, Saragliadis A, Hiruma Y, Overhand M, Ubbink M (2008) Design, synthesis, and evaluation of a lanthanide chelating protein probe: CLaNP-5 yields predictable paramagnetic effects independent of environment. J Am Chem Soc 130:14802–14812
Liu WM, Overhand M, Ubbink M (2014) The application of paramagnetic lanthanoid ions in NMR spectroscopy on proteins. Coord Chem Rev 273–274:2–12
Loh CT, Ozawa K, Tuck KL, Barlow N, Huber T, Otting G, Graham B (2013) Lanthanide tags for site-specific ligation to an unnatural amino acid and generation of pseudocontact shifts in proteins. Bioconj Chem 24:260–268
Mironov VS, Galyametdinov YG, Ceulemans A, Görller-Walrand C, Binnemans K (2001) Influence of crystal-field perturbations on the room-temperature magnetic anisotropy of lanthanide complexes. Chem Phys Lett 345:132–140
Müntener T, Häussinger D, Selenko P, Theillet FX (2016) In-cell protein structures from 2D NMR experiments. J Phys Chem Lett 7:2821–2825
Natrajan LS, Khoabane NM, Dadds BL, Muryn CA, Pritchard RG, Heath SL, Kenwright AM, Kuprov I, Faulkner S (2010) Probing the structure, conformation, and stereochemical exchange in a family of lanthanide complexes derived from tetrapyridyl-appended cyclen. Inorg Chem 49:7700–7709
Neese F (2012) The ORCA program system. Wiley Interdiscip Rev Comput Mol Sci 2:73–78
Nitsche C, Otting G (2017) Pseudocontact shifts in biomolecular NMR using paramagnetic metal tags. Prog Nucl Magn Reson Spectrosc 98–99:20–49
Opina ACL, Strickland M, Lee YS, Tjandra N, Byrd RA, Swenson RE, Vasalatiy O (2016) Analysis of the isomer ratios of polymethylated-DOTA complexes and the implications on protein structural studies. Dalton Trans 45:4673–4687
Otting G (2010) Protein NMR using paramagnetic ions. Annu Rev Biophys 39:387–405
Pan BB, Yang F, Ye Y, Wu Q, Li C, Huber T, Su XC (2016) 3D structure determination of a protein in living cells using paramagnetic NMR spectroscopy. Chem Commun 52:10237–10240
Parker D, Dickins RS, Puschmann H, Crossland C, Howard JAK (2002) Being excited by lanthanide coordination complexes: aqua species, chirality, excited-state chemistry, and exchange dynamics. Chem Rev 102:1977–2010
Perdew JP (1986) Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys Rev B 33:8822–8824
Peters F, Maestre-Martinez M, Leonov A, Kovacic L, Becker S, Boelens R, Griesinger C (2011) Cys-Ph-TAHA: a lanthanide binding tag for RDC and PCS enhanced protein NMR. J Biomol NMR 51:329–337
Pintacuda G, Park AY, Keniry MA, Dixon NE, Otting G (2006) Lanthanide labeling offers fast NMR approach to 3D structure determinations of protein-protein complexes. J Am Chem Soc 128:3696–3702
Ramage R, Green J, Muir TW, Ogunjobi OM, Love S, Shaw K (1994) Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem J 299:151–158
Ranganathan RS, Pillai RK, Raju N, Fan H, Nguyen H, Tweedle MF, Desreux JF, Jacques V (2002a) Polymethylated DOTA ligands. 1. Synthesis of rigidified ligands and studies on the effects of alkyl substitution on acid-base properties and conformational mobility. Inorg Chem 41:6846–6855
Ranganathan RS, Raju N, Fan H, Zhang X, Tweedle MF, Desreux JF, Jacques V (2002b) Polymethylated DOTA ligands. 2. Synthesis of rigidified lanthanide chelates and studies on the effect of alkyl substitution on conformational mobility and relaxivity. Inorg Chem 41:6856–6866
Redfern PC, Zapol P, Curtiss LA, Raghavachari K (2000) Assessment of Gaussian-3 and density functional theories for enthalpies of formation of C1-C16 alkanes. J Phys Chem A 104:5850–5854
Regueiro-Figueroa M, Bensenane B, Ruscsák E, Esteban-Gómez D, Charbonnière LJ, Tircsó G, Tóth I, Blas AD, Rodríguez-Blas T, Platas-Iglesias C (2011) Lanthanide DOTA-like complexes containing a picolinate pendant: structural entry for the design of LnIII-based luminescent probes. Inorg Chem 50:4125–4141
Sass J, Cordier F, Hoffmann A, Rogowski M, Cousin A, Omichinski JG, Löwen H, Grzesiek S (1999) Purple membrane induced alignment of biological macromolecules in the magnetic field. J Am Chem Soc 121:2047–2055
Schmitz C, Stanton-Cook MJ, Su X-C, Otting G, Huber T (2008) Numbat: an interactive software tool for fitting ∆χ-tensors to molecular coordinates using pseudocontact shifts. J Biomol NMR 41:179–189
Shannon RD (1976) Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Cryst A 32:751–767
Shishmarev D, Otting G (2013) How reliable are pseudocontact shifts induced in proteins and ligands by mobile paramagnetic metal tags? A modelling study. J Biomol NMR 56:203–216
Sousa SF, Fernandes PA, Ramos MJ (2007) General performance of density functionals. J Phys Chem A 111:10439–10452
Strickland M, Schwieters CD, Göbl C, Opina ACL, Strub MP, Swenson RE, Vasalatiy O, Tjandra N (2016) Characterizing the magnetic susceptibility tensor of lanthanide-containing polymethylated-DOTA complexes. J Biomol NMR 66:125–139
Su XC, Man B, Beeren S, Liang H, Simonsen S, Schmitz C, Huber T, Messerle BA, Otting G (2008) A dipicolinic acid tag for rigid lanthanide tagging of proteins and paramagnetic NMR spectroscopy. J Am Chem Soc 130:10486–10487
Su XC, Liang H, Loscha KV, Otting G (2009) [Ln(DPA)3]3– is a convenient paramagnetic shift reagent for protein NMR studies. J Am Chem Soc 131:10352–10353
Suturina EA, Mason K, Geraldes C, Kuprov I, Parker D (2017) Beyond Bleaney’s theory: experimental and theoretical analysis of periodic trends in lanthanide-induced chemical shift. Angew Chem Int Ed 56:12215–12218
Takano Y, Houk KN (2005) Benchmarking the conductor-like polarizable continuum model (CPCM) for aqueous solvation free energies of neutral and ionic organic molecules. J Chem Theory Comput 1:70–77
Varghese S, Halling PJ, Häussinger D, Wimperis S (2016) High-resolution structural characterization of a heterogeneous biocatalyst using solid-state NMR. J Phys Chem C 120:28717–28726
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct, Funct, Bioinform 59:687–696
Wöhnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H (2003) Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J Am Chem Soc 125:13338–13339
Acknowledgements
The Chemistry Department of the University of Basel and the Swiss National Science Foundation Grant 200021_130263 are acknowledged for financial support. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing core facility at University of Basel. Biological structures were generated using the open source software PyMOL (http://www.pymol.org/). C.E. Housecroft, E.C. Constable, T. Müntener and R. Vogel are acknowledged for helpful discussions. R.A. Byrd is gratefully acknowledged for a gift of M4-cyclen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Joss, D., Walliser, R.M., Zimmermann, K. et al. Conformationally locked lanthanide chelating tags for convenient pseudocontact shift protein nuclear magnetic resonance spectroscopy. J Biomol NMR 72, 29–38 (2018). https://doi.org/10.1007/s10858-018-0203-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-018-0203-4