Abstract
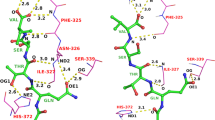
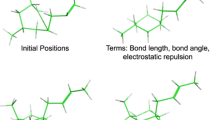
An algorithm, CYLIB, is presented for converting molecular topology descriptions from the PDB Chemical Component Dictionary into CYANA residue library entries. The CYANA structure calculation algorithm uses torsion angle molecular dynamics for the efficient computation of three-dimensional structures from NMR-derived restraints. For this, the molecules have to be represented in torsion angle space with rotations around covalent single bonds as the only degrees of freedom. The molecule must be given a tree structure of torsion angles connecting rigid units composed of one or several atoms with fixed relative positions. Setting up CYANA residue library entries therefore involves, besides straightforward format conversion, the non-trivial step of defining a suitable tree structure of torsion angles, and to re-order the atoms in a way that is compatible with this tree structure. This can be done manually for small numbers of ligands but the process is time-consuming and error-prone. An automated method is necessary in order to handle the large number of different potential ligand molecules to be studied in drug design projects. Here, we present an algorithm for this purpose, and show that CYANA structure calculations can be performed with almost all small molecule ligands and non-standard amino acid residues in the PDB Chemical Component Dictionary.









Similar content being viewed by others
References
Alipanahi B, Gao X, Karakoc E, Donaldson L, Li M (2009) PICKY: a novel SVD-based NMR spectra peak picking method. Bioinformatics 25:i268–i275
Allen FH, Bellard S, Brice MD, Cartwright BA, Doubleday A, Higgs H, Hummelink T, Hummelink-Peters BG, Kennard O, Motherwell WDS, Rodgers JR, Watson DG (1979) The Cambridge crystallographic data centre: computer-based search, retrieval, analysis and display of information. Acta Crystallogr Sect B Struct Commun 35:2331–2339
Arkin MR, Wells JA (2004) Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov 3:301–317
Bahrami A, Assadi AH, Markley JL, Eghbalnia HR (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comp Biol 5:e1000307
Bardiaux B, Malliavin T, Nilges M (2012) ARIA for solution and solid-state NMR. Meth Mol Biol 831:453–483
Bartels C, Güntert P, Billeter M, Wüthrich K (1997) GARANT—a general algorithm for resonance assignment of multidimensional nuclear magnetic resonance spectra. J Comput Chem 18:139–149
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucl Acids Res 28:235–242
Bourne PE, Berman HM, McMahon B, Watenpaugh KD, Westbrook JD, Fitzgerald PMD (1997) Macromolecular crystallographic information file. Methods Enzymol 277:571–590
Cormen TH, Leiserson CE, Rivest RL (1990) Introduction to algorithms. MIT Press, Cambridge
Guerry P, Herrmann T (2011) Advances in automated NMR protein structure determination. Q Rev Biophys 44:257–309
Güntert P (2009) Automated structure determination from NMR spectra. Eur Biophys J 38:129–143
Güntert P, Buchner L (2015) Combined automated NOE assignment and structure calculation with CYANA. J Biomol NMR. doi:10.1007/s10858-015-9924-9
Güntert P, Braun W, Wüthrich K (1991) Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol 217:517–530
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298
Hamada T, Matsunaga S, Fujiwara M, Fujita K, Hirota H, Schmucki R, Güntert P, Fusetani N (2010) Solution structure of polytheonamide B, a highly cytotoxic non-ribosomal polypeptide from marine sponge. J Am Chem Soc 132:12941–12945
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227
Huang YJ, Tejero R, Powers R, Montelione GT (2006) A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins 62:587–603
Ingram RN, Orth P, Strickland CL, Le HV, Madison V, Beyer BM (2006) Stabilization of the autoproteolysis of TNF-alpha converting enzyme (TACE) results in a novel crystal form suitable for structure-based drug design studies. Protein Eng Des Sel 19:155–161
Jain A, Vaidehi N, Rodriguez G (1993) A fast recursive algorithm for molecular dynamics simulation. J Comput Phys 106:258–268
Kallen J, Spitzfaden C, Zurini MGM, Wider G, Widmer H, Wüthrich K, Walkinshaw MD (1991) Structure of human cyclophilin and its binding site for cyclosporin A determined by X-ray crystallography and NMR spectroscopy. Nature 353:276–279
Klukowski P, Walczak MJ, Gonczarek A, Boudet J, Wider G (2015) Computer vision—based automated peak picking applied to protein NMR spectra. Bioinformatics. doi:10.1093/bioinformatics/btv318
Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55
Koradi R, Billeter M, Engeli M, Güntert P, Wüthrich K (1998) Automated peak picking and peak integration in macromolecular NMR spectra using AUTOPSY. J Magn Reson 135:288–297
López-Méndez B, Güntert P (2006) Automated protein structure determination from NMR spectra. J Am Chem Soc 128:13112–13122
Sadowski J, Gasteiger J, Klebe G (1994) Comparison of automatic three-dimensional model builders using 639 X-ray structures. J Chem Inf Comput Sci 34:1000–1008
Schmidt E, Güntert P (2012) A new algorithm for reliable and general NMR resonance assignment. J Am Chem Soc 134:12817–12829
Shuker SB, Hajduk PJ, Meadows RP, Fesik SW (1996) Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531–1534
Weber C, Wider G, von Freyberg B, Traber R, Braun W, Widmer H, Wüthrich K (1991) NMR structure of cyclosporin A bound to cyclophilin in aqueous solution. Biochemistry 30:6563–6574
Westbrook JD, Shao C, Feng Z, Zhuravleva M, Velankar S, Young J (2015) The chemical component dictionary: complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the Protein Data Bank. Bioinformatics 31:1274–1278
Wüthrich K (1986) NMR of proteins and nucleic acids. Wiley, New York
Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S (2008) Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol 378:634–652
Acknowledgments
We gratefully acknowledge financial support by the Lichtenberg program of the Volkswagen Foundation and a Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yilmaz, E.M., Güntert, P. NMR structure calculation for all small molecule ligands and non-standard residues from the PDB Chemical Component Dictionary. J Biomol NMR 63, 21–37 (2015). https://doi.org/10.1007/s10858-015-9959-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-9959-y