Abstract
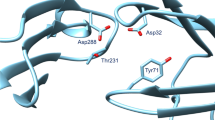
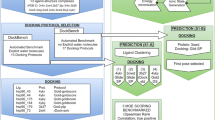
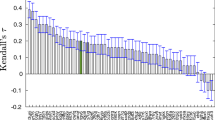
In this paper we describe our approaches to predict the binding mode of twenty BACE1 ligands as part of Grand Challenge 4 (GC4), organized by the Drug Design Data Resource. Calculations for all submissions (except for one, which used AutoDock4.2) were performed using AutoDock-GPU, the new GPU-accelerated version of AutoDock4 implemented in OpenCL, which features a gradient-based local search. The pose prediction challenge was organized in two stages. In Stage 1a, the protein conformations associated with each of the ligands were undisclosed, so we docked each ligand to a set of eleven receptor conformations, chosen to maximize the diversity of binding pocket topography. Protein conformations were made available in Stage 1b, making it a re-docking task. For all calculations, macrocyclic conformations were sampled on the fly during docking, taking the target structure into account. To leverage information from existing structures containing BACE1 bound to ligands available in the PDB, we tested biased docking and pose filter protocols to facilitate poses resembling those experimentally determined. Both pose filters and biased docking resulted in more accurate docked poses, enabling us to predict for both Stages 1a and 1b ligand poses within 2 Å RMSD from the crystallographic pose. Nevertheless, many of the ligands could be correctly docked without using existing structural information, demonstrating the usefulness of physics-based scoring functions, such as the one used in AutoDock4, for structure based drug design.




Similar content being viewed by others
References
Wagner J, Churas C, Liu S, Swift R, Chiu M, Shao C, Feher V, Burley S, Gilson M, Amaro R (2018) bioRxiv
Gaieb Z, Parks CD, Chiu M, Yang H, Shao C, Walters WP, Lambert MH, Nevins N, Bembenek SD, Ameriks MK et al (2019) J Comput-Aided Mol Des 33(1):1
Gaieb Z, Liu S, Gathiaka S, Chiu M, Yang H, Shao C, Feher VA, Walters WP, Kuhn B, Rudolph MG et al (2018) J Comput-Aided Mol Des 32(1):1
Gathiaka S, Liu S, Chiu M, Yang H, Stuckey JA, Kang YN, Delproposto J, Kubish G, Dunbar JB, Carlson HA et al (2016) J Comput-Aided Mol Des 30(9):651
Smith RD, Damm-Ganamet KL, Dunbar JB Jr, Ahmed A, Chinnaswamy K, Delproposto JE, Kubish GM, Tinberg CE, Khare SD, Dou J et al (2015) J Chem Inf Model 56(6):1022
Yin J, Henriksen NM, Slochower DR, Shirts MR, Chiu MW, Mobley DL, Gilson MK (2017) J Comput-Aided Mol Des 31(1):1
Rizzi A, Murkli S, McNeill JN, Yao W, Sullivan M, Gilson MK, Chiu MW, Isaacs L, Gibb BC, Mobley DL et al (2018) J Comput-Aided Mol Des 32(10):937
Bannan CC, Burley KH, Chiu M, Shirts MR, Gilson MK, Mobley DL (2016) J Comput-Aided Mol Des 30(11):927
Llinas A, Avdeef A (2019) J Chem Inf Model 59(6):3036
Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R et al (2016) Proteins 84:323
Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A (2018) Proteins 86:7
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) J Nucleic Acids Res 28(1):235
Santos-Martins D, Solis-Vasquez L, Koch A, Forli S (2019). https://doi.org/10.26434/chemrxiv.9702389.v1
Forli S, Botta M (2007) J Chem Inf Model 47(4):1481
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) J Cheminform 3(1):33
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) J Comput Chem 28(6):1145
Xu Y, Li Mj, Greenblatt H, Chen W, Paz A, Dym O, Peleg Y, Chen T, Shen ,X, He J et al (2012) Acta Crystallogr Sect D 68(1):13
DeLano WL (2002) CCP4 Newsl Protein Crystallogr 40(1):82
Word JM, Lovell SC, Richardson JS, Richardson DC (1999) J Mol Biol 285(4):1735
Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ (2016) Nat Protoc 11(5):905
Forli S, Olson AJ (2012) J Med Chem 55(2):623
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) J Comput Chem 19(14):1639
Solis FJ, Wets RJB (1981) Math Oper Res 6(1):19
Zeiler MD (2012) arXiv preprint arXiv:1212.5701
Borg I, Groenen P (2003) J Educ Meas 40(3):277–280. https://doi.org/10.1111/j.1745-3984.2003.tb01108.x
Jolliffe I (2011) Principal component analysis. Springer, New York
Sittel F, Jain A, Stock G (2014) J Chem Phys 141(1):07B605
Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O (2011) J Comput Chem 32(10):2319
Gowers RJ, Linke M, Barnoud J, Reddy TJ, Melo MN, Seyler SL, Domański J, Dotson DL, Buchoux S, Kenney IM et al (2016) In: Proceedings of the 15th python in science conference, vol 98. SciPy Austin, TX
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V et al (2011) J Mach Learn Res 12(Oct):2825
De Leeuw J (2011) Applications of Convex Analysis to Multidimensional Scaling. UCLA: Department of Statistics, UCLA. Retrieved from https://escholarship.org/uc/item/4ps3b5mj
De Silva V, Tenenbaum JB (2004) Sparse multidimensional scaling using landmark points. Tech. rep., Technical report, Stanford University
Patel D, Antwi J, Koneru PC, Serrao E, Forli S, Kessl JJ, Feng L, Deng N, Levy RM, Fuchs JR et al (2016) J Biol Chem 291(45):23569
Xu S, Hermanson DJ, Banerjee S, Ghebreselasie K, Clayton GM, Garavito RM, Marnett LJ (2014) J Biol Chem 289(10):6799
Fu H, Cui M, Zhao L, Tu P, Zhou K, Dai J, Liu B (2015) J Med Chem 58(17):6972
Acknowledgements
This work was supported by the National Institutes of Health R01-GM069832 (DSM, JE, SF), and U54-GM103368 (GB). LSV and AK thank the German Academic Exchange Service (DAAD) and the Peruvian National Program for Scholarships and Educational Loans (PRONABEC) for financial aid.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Santos-Martins, D., Eberhardt, J., Bianco, G. et al. D3R Grand Challenge 4: prospective pose prediction of BACE1 ligands with AutoDock-GPU. J Comput Aided Mol Des 33, 1071–1081 (2019). https://doi.org/10.1007/s10822-019-00241-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00241-9