Abstract
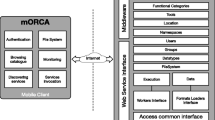
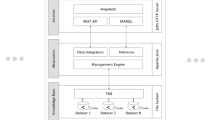
Biomedical researchers can leverage Grid computing technology to address their increasing demands for data- and compute-intensive data analysis. However, usage of existing Grid infrastructures remains difficult for them. The e-infrastructure for biomedical science (e-BioInfra) is a platform with services that shield middleware complexities, in particular workflow management and monitoring. These services can be invoked from a web-based interface, called e-BioInfra Gateway, to perform large scale data analysis experiments, such that the biomedical researchers can focus on their own research problems. The gateway was designed to simplify usage both by biomedical researchers and e-BioInfra administrators, and to support straightforward extensions with new data analysis methods. In this paper we present the architecture and implementation of the gateway, also showing statistics for its usage. We also share lessons learned during the gateway development and operation. The gateway is currently used in several biomedical research projects and in teaching medical students the principles of data analysis.
Similar content being viewed by others
References
Alfieri, R., Cecchini, R., Ciaschini, V., dell’Agnello, L., Frohner, Á., Gianoli, A., Lõrentey, K., Spataro, F.: Voms, an authorization system for virtual organizations. In: Fernández Rivera, F., Bubak, M., Gómez Tato, A., Doallo, R. (eds.) Grid Computing. Lecture Notes in Computer Science, vol. 2970, pp. 33–40. Springer, Berlin/Heidelberg (2004)
Altunay, M., Avery, P., Blackburn, K., Bockelman, B., Ernst, M., Fraser, D., Quick, R., Gardner, R., Goasguen, S., Levshina, T., Livny, M., McGee, J., Olson, D., Pordes, R., Potekhin, M., Rana, A., Roy, A., Sehgal, C., Sfiligoi, I., Wuerthwein, F.: A Science Driven Production Cyberinfrastructure—the Open Science Grid. J. Grid Computing 9, 201–218 (2011)
Andronico, G., Ardizzone, V., Barbera, R., Becker, B., Bruno, R., Calanducci, A., Carvalho, D., Ciuffo, L., Fargetta, M., Giorgio, E., La Rocca, G., Masoni, A., Paganoni, M., Ruggieri, F., Scardaci, D.: e-infrastructures for e-science: a global view. J. Grid Computing 9, 155–184 (2011)
Barbera, R., Andronico, G., Donvito, G., Falzone, A., Keijser, J.J., Rocca, G.L., Milanesi, L., Maggi, G.P., Vicario, S.: A Grid portal with robot certificates for bioinformatics phylogenetic analyses. Concurrency Computat.: Pract. Exper. 23(3), 246–255 (2011)
Berkeley Database Information Index (BDII): https://twiki.cern.ch/twiki/bin/view/EGEE/BDII. Accessed 23 May 2012
Basney, J., Humphrey, M., Welch, V.: The myproxy online credential repository. Softw. Pract. Exper. 35(9), 801–816 (2005)
Bertini, I., Case, D.A., Ferella, L., Giachetti, A., Rosato, A.: A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics 27(17), 2384–2390 (2011). doi:10.1093/bioinformatics/btr415
Birkenheuer, G., Blunk, D., Breuers, S., Brinkmann, A., Fles, G., Gesing, S., et al.: MoSGrid: progress of workflow driven chemical simulations. In: Proceedings of Grid Workflow Workshop (GWW) (2011)
Breton, V., Dean, K., Solomonides, T., Blanquer, I., Hernandez, V., Medico, E., Maglaveras, N., Benkner, S., Lonsdale, G., Lloyd, S., Hassan, K., McClatchey, R., Miguet, S., Montagnat, J., Pennec, X., De Neve, W., De Wagter, C., Heeren, G., Maigne, L., Nozaki, K., Taillet, M., Bilofsky, H., Ziegler, R., Hoffman, M., Jones, C., Cannataro, M., Veltri, P., Aloisio, G., Fiore, S., Mirto, M., Chouvarda, I., Koutkias, V., Malousi, A., Lopez, V., Oliveira, I., Sanchez, J.P., Martin-Sanchez, F., De Moor, G., Claerhout, B., Herveg, J.A.: The healthgrid white paper. Stud. Health Technol. Inform. 112, 249–321 (2005)
Caan, M., Shahand, S., Vos, F., van Kampen, A., Olabarriaga, S.: Evolution of Grid-based services for diffusion tensor image analysis. Future Gener. Comput. Syst. 28(8), 1194–1204 (2012)
Caan, M., Vos, F., van Kampen, A., Olabarriaga, S., van Vliet, L.: Gridifying a diffusion tensor imaging analysis pipeline. In: 10th IEEE/ACM International Conference on Cluster, Cloud and Grid Computing (CCGrid), pp. 733–738 (2010)
Camarasu-Pop, S., Glatard, T., Moscicki, J.T., Benoit-Cattin, H., Sarrut, D.: Dynamic partitioning of GATE Monte-Carlo simulations on EGEE. J. Grid Computing 8(2), 241–259 (2010)
Casajus, A., Graciani, R., Paterson, S., Tsaregorodtsev, A., the Lhcb Dirac Team: Dirac pilot framework and the dirac workload management system. J. Phys.: Conf. Ser. 219(6), 062,049 (2010)
DTI Preprocessing on the e-BioinfraGateway: http://www.bioinformaticslaboratory.nl/twiki/bin/view/EBioScience/PredtiUserDoc. Accessed 23 May 2012
EGI Science Gateways: http://www.egi.eu/services/support/science-gateways/index.html. Accessed 23 May 2012
Ferrari, T., Gaido, L.: Resources and services of the EGEE production infrastructure. J. Grid Computing 9, 119–133 (2011)
Ferreira da Silva, R., Camarasu-Pop, S., Grenier, B., Hamar, V., Manset, D., Montagnat, J., Revillard, J., Balderrama, J.R., Tsaregorodtsev, A., Glatard, T.: Multi-infrastructure workflow execution for medical simulation in the Virtual Imaging Platform. In: Proceedings of HealthGrid 2011. Bristol, UK (2011)
Fischl, B., van der Kouwe, A., Destrieux, C., Halgren, E., Ségonne, F., Salat, D.H., Busa, E., Seidman, L.J., Goldstein, J., Kennedy, D., Caviness, V., Makris, N., Rosen, B., Dale, A.M.: Automatically parcellating the human cerebral cortex. Cereb. Cortex 14(1), 11–22 (2004)
FMRIB’s Diffusion Toolbox—BEDPOSTX: http://www.fmrib.ox.ac.uk/fsl/fdt/fdt_bedpostx.html. Accessed 23 May 2012
Genome Compare on the e-BioinfraGateway: http://www.bioinformaticslaboratory.nl/twiki/bin/view/EBioScience/GenomeCompareUserDoc. Accessed 23 May 2012
Gesing, S., Hemert, J.v., Kacsuk, P., Kohlbacher, O.: Special issue: portals for life sciences—providing intuitive access to bioinformatic tools. Concurrency Computat.: Pract. Exper. 23(3), 223–234 (2011)
Glatard, T., Montagnat, J., Lingrand, D., Pennec, X.: Flexible and efficient workflow deployment of data-intensive applications on Grids with MOTEUR. Int. J. High Perform. Comput. Appl. 22(3), 347–360 (2008)
Goodale, T., Jha, S., Kaiser, H., Kielmann, T., Kleijer, P., Von Laszewski, G., Lee, C., Merzky, A., Rajic, H., Shalf, J.: Saga: a simple api for Grid applications. High-level application programming on the Grid. Comput. Methods Sci. Technol. 12(1), 7–20 (2006)
Helmer, K.G., Ambite, J.L., Ames, J., Ananthakrishnan, R., Burns, G., Chervenak, A.L., Foster, I., Liming, L., Keator, D., Macciardi, F., Madduri, R., Navarro, J.P., Potkin, S., Rosen, B., Ruffins, S., Schuler, R., Turner, J.A., Toga, A., Williams, C., Kesselman, C., for the Biomedical Informatics Research Network: Enabling collaborative research using the Biomedical Informatics Research Network (BIRN). J. Am. Med. Inform. Assoc. 18(4), 416–422 (2011)
Hey, T., Tansley, S., Tolle, K. (eds.): The Fourth Paradigm: Data-Intensive Scientific Discovery. Microsoft Research (2009)
Kacsuk, P.: P-GRADE portal family for Grid infrastructures. Concurrency Computat.: Pract. Exper. 23(3), 235–245 (2011)
Kim, J., Maddineni, S., Jha, S.: Building gateways for life-science applications using the dynamic application runtime environment (dare) framework. In: Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, TG ’11, pp. 38:1–38:8. ACM, New York (2011)
Kiss, T., Greenwell, P., Heindl, H., Terstyanszky, G., Weingarten, N.: Parameter sweep workflows for modelling carbohydrate recognition. J. Grid Computing 8, 587–601 (2010)
Klarenbeek, P.L., Tak, P.P., van Schaik, B.D.C., Zwinderman, A.H., Jakobs, M.E., Zhang, Z., van Kampen, A.H.C., van Lier, R.A.W., Baas, F., de Vries, N.: Human T-cell memory consists mainly of unexpanded clones. Immunol. Lett. 133(1), 42–48 (2010)
Korkhov, V., Krefting, D., Kukla, T., Terstyanszky, G.Z., Caan, M., Olabarriaga, S.D.: Exploring workflow interoperability tools for neuroimaging data analysis. In: Proceedings of the 6th Workshop on Workflows in Support of Large-Scale Science, WORKS ’11, pp. 87–96. ACM, New York (2011)
Krefting, D., Bart, J., Beronov, K., Dzhimova, O., Falkner, J., Hartung, M., Hoheisel, A., Knoch, T.A., Lingner, T., Mohammed, Y., Peter, K., Rahm, E., Sax, U., Sommerfeld, D., Steinke, T., Tolxdorff, T., Vossberg, M., Viezens, F., Weisbecker, A.: MediGRID: Towards a user friendly secured Grid infrastructure. Future Gener. Comput. Syst. 25(3), 326–336 (2009)
Luyf, A., van Schaik, B., de Vries, M., Baas, F., van Kampen, A., Olabarriaga, S.: Initial steps towards a production platform for DNA sequence analysis on the Grid. BMC Bioinformatics 11(1), 598 (2010)
Marco, C., Fabio, C., Alvise, D., Antonia, G., Francesco, G., Alessandro, M., Moreno, M., Salvatore, M., Fabrizio, P., Luca, P., Francesco, P.: The glite workload management system. In: Abdennadher, N., Petcu, D. (eds.) Advances in Grid and Pervasive Computing. Lecture Notes in Computer Science, vol. 5529, pp. 256–268. Springer, Berlin (2009)
Model–view–controller—Wikipedia: http://en.wikipedia.org/wiki/Model-view-controller. Accessed 23 May 2012
Montagnat, J., Isnard, B., Glatard, T., Maheshwari, K., Fornarino, M.: A data-driven workflow language for Grids based on array programming principles. In: Proceedings of the 4th Workshop on Workflows in Support of Large-Scale Science (WORKS) (2009)
Moscicki, J.T., Lamanna, M., Bubak, M., Sloot, P.M.A.: Processing moldable tasks on the Grid: late job binding with lightweight user-level overlay. Future Gener. Comput. Syst. 27(6), 725–736 (2011)
Novotny, J., Russell, M., Wehrens, O.: GridSphere: a portal framework for building collaborations. Concurrency Computat.: Pract. Exper. 16(5), 503–513 (2004)
Olabarriaga, S.D., Glatard, T., de Boer, P.T.: A virtual laboratory for medical image analysis. IEEE Trans. Inf. Technol. Biomed. 14(4), 979–985 (2010)
Olabarriaga, S.D., Glatard, T., Boulebiar, K., de Boer, P.T.: From “low hanging” to “user ready”: initial steps into a HealthGrid. In: Global Healthgrid: e-Science Meets Biomedical Informatics—Proceedings of HealthGrid 2008, vol. 138, pp. 70–79 (2008)
Pandey, S., Voorsluys, W., Rahman, M., Buyya, R., Dobson, J.E., Chiu, K.: A Grid workflow environment for brain imaging analysis on distributed systems. Concurrency Computat.: Pract. Exper. 21(16), 2118–2139 (2009)
Peters, B.D., Machielsen, M.W.J., Hoen, W.P., Caan, M.W.A., Malhotra, A.K., Szeszko, P.R., Duran, M., Olabarriaga, S.D., de Haan, L.: Polyunsaturated fatty acid concentration predicts myelin integrity in earlyphase psychosis. Schizophr. Bull. (2012). doi:10.1093/schbul/sbs089
Redolfi, A., McClatchey, R., Anjum, A., Zijdenbos, A., Manset, D., Barkhof, F., Spenger, C., Legré, Y., Wahlund, L.O., di San Pietro, C.B., Frisoni, G.B.: Grid infrastructures for computational neuroscience: the neuGRID example. Future Neurol. 4(6), 703–722 (2009)
Shahand, S., Caan, M., van Kampen, A., Olabarriaga, S.: Integrated support for neuroscience research: from study design to publication. In: Proceedings of HealthGrid 2012. Amsterdam, NL (2012)
Shahand, S., Santcroos, M., Mohammed, Y., Korkhov, V., Luyf, A., van Kampen, A., Olabarriaga, S.: Front-ends to biomedical data analysis on Grids. In: Proceedings of HealthGrid 2011. Bristol, UK (2011)
Stewart, G.A., Cameron, D., Cowan, G.A., McCance, G.: Storage and data management in egee. In: Proceedings of the fifth Australasian symposium on ACSW frontiers, vol. 68, ACSW ’07, pp. 69–77. Australian Computer Society, Inc., Darlinghurst, Australia (2007)
The BigGrid Project: http://www.biggrid.nl. Accessed 23 May 2012
The Engineframe Project: http://www.enginframe.com. Accessed 23 May 2012
The gLite Project: http://glite.cern.ch. Accessed 23 May 2012
The Google Web Toolkit. https://developers.google.com/web-toolkit. Accessed 23 May 2012
The Hibernate Project: http://www.hibernate.org. Accessed 23 May 2012
The Liferay Project: http://www.liferay.com. Accessed 23 May 2012
The Pylons Project: http://www.pylonsproject.org. Accessed 23 May 2012
The Spring Project: http://www.springsource.org. Accessed 23 May 2012
Using an Aladdin eToken PRO to store Grid certificates: http://www.nikhef.nl/pub/projects/grid/gridwiki/index.php/EToken. Accessed 23 May 2012
van Wingen, G.A., Geuze, E., Caan, M.W.A., Kozicz, T., Olabarriaga, S.D., Denys, D., Vermetten, E., Fernández, G.: Persistent and reversible consequences of combat stress on the mesofrontal circuit and cognition. Proc. Natl. Acad. Sci. (PNAS) (2012). doi:10.1073/pnas.1206330109
Wilkins-Diehr, N., Gannon, D., Klimeck, G., Oster, S., Pamidighantam, S.: TeraGrid science gateways and their impact on science. Comput. 41(11), 32 –41 (2008)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shahand, S., Santcroos, M., van Kampen, A.H.C. et al. A Grid-Enabled Gateway for Biomedical Data Analysis. J Grid Computing 10, 725–742 (2012). https://doi.org/10.1007/s10723-012-9233-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10723-012-9233-4